6F3W
 
 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F46
 
 | |
6F4Z
 
 | |
6F55
 
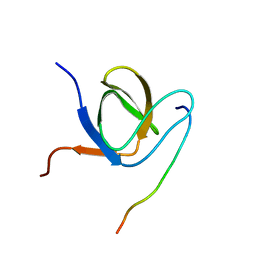 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
6F61
 
 | |
6F7E
 
 | |
6F7M
 
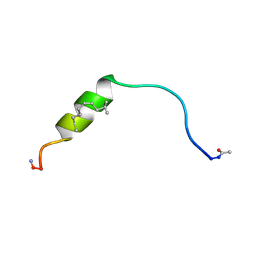 | | NMR structure of EphA2-Sam stapled peptides (S13ST) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6F7N
 
 | | NMR structure of EphA2-Sam stapled peptides (S13STshort) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6F7O
 
 | | NMR structure of an Odin-Sam1 stapled peptide | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Leone, M, Mercurio, F.A. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6F8E
 
 | | PH domain from TgAPH | | Descriptor: | Pleckstrin homology domain | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Phosphatidic Acid Sensing by APH in Apicomplexan Parasites.
Structure, 26, 2018
|
|
6F98
 
 | | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1
To Be Published
|
|
6F99
 
 | |
6F9A
 
 | | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5 | | Descriptor: | ER quality-control lectin | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5
To Be Published
|
|
6FBL
 
 | | NMR Solution Structure of MINA-1(254-334) | | Descriptor: | MINA-1 | | Authors: | Michel, E, Allain, F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans.
Cell Death Differ., 26, 2019
|
|
6FC9
 
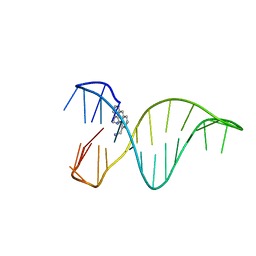 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
6FCE
 
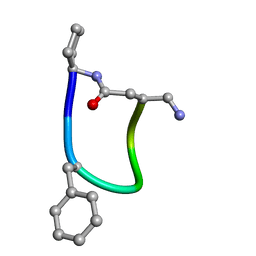 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
6FD7
 
 | |
6FDP
 
 | |
6FDT
 
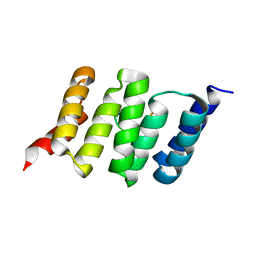 | |
6FE6
 
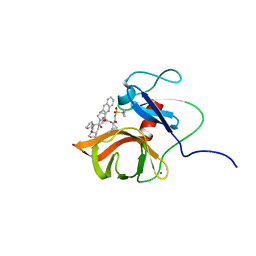 | | Solution structure of a last generation P2-P4 macrocyclic inhibitor | | Descriptor: | (3aR,7S,10S,12R,24aR)-7-cyclopentyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-5,8-dioxo-1,2,3,3a,5,6,7,8,11,12,20,21,22,23,24,24a-hexadecahydro-10H-9,12-methanocyclopenta[18,19][1,10,3,6]dioxadiazacyclononadecino[12,11-b]quinoline-10-carboxamide, Non-structural 3 protease, ZINC ION | | Authors: | Gallo, M, Eliseo, T, Cicero, D.O. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a last generation macrocyclic inhibitor. Hepatitis C virus NS3 protease complex: when S prime region occupancy is not enough to stabilize the protein conformation in the absence of NS4A.
To Be Published
|
|
6FEG
 
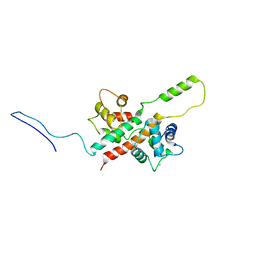 | | Solution Structure of CaM/Kv7.2-hAB Complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2,Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Bernardo-Seisdedos, G, Villarroel, A, Millet, O. | | Deposit date: | 2018-01-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and energy landscape for the Ca2+gating and calmodulation of the Kv7.2 K+channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FEH
 
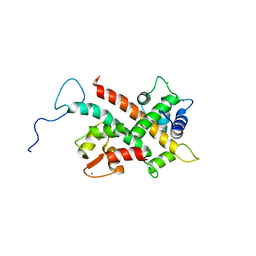 | | Solution Structure of CaM/Kv7.2-hAB Complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2,Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Bernardo-Seisdedos, G, Villarroel, A, Millet, O. | | Deposit date: | 2018-01-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis and energy landscape for the Ca2+gating and calmodulation of the Kv7.2 K+channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FFQ
 
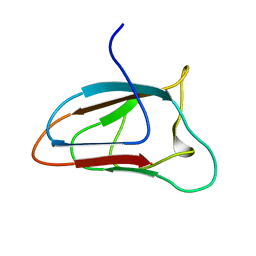 | | Solution NMR structure of CBM64 from S.thermophila | | Descriptor: | Glycosyl hydrolase family 5 cellulase CBM64 | | Authors: | Heikkinen, H.A, Iwai, H. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13 C- and Uniformly 100% 15 N-Labeled Sample.
Molecules, 26, 2021
|
|
