2MNU
 
 | |
2MNW
 
 | |
2MNX
 
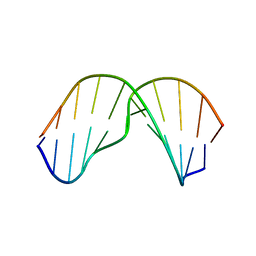 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | Descriptor: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2MO0
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp | | Descriptor: | Cold-shock DNA-binding domain protein | | Authors: | Jin, B, Jeong, K.W, Kim, Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
2MO1
 
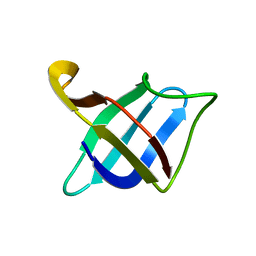 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp with dT7 | | Descriptor: | Cold-shock DNA-binding domain protein | | Authors: | Jin, B, Jeong, K.W, Kim, Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
2MO2
 
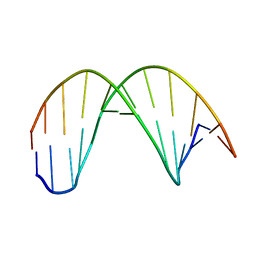 | |
2MO5
 
 | | hIFABP-oleate complex | | Descriptor: | Fatty acid-binding protein, intestinal, OLEIC ACID | | Authors: | Patil, R, Mohanty, B, Headey, S, Porter, C, Scanlon, M. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of oleate bound human intestinal fatty acid binding protein
To be Published
|
|
2MO7
 
 | |
2MOA
 
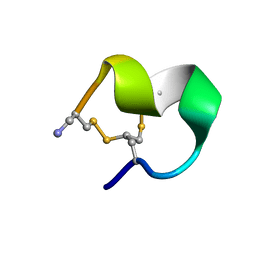 | |
2MOC
 
 | |
2MOE
 
 | |
2MOF
 
 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | Descriptor: | Bacterial Ig-like domain, group 2 | | Authors: | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
2MOI
 
 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOK
 
 | | holo_FldA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2014-04-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR study of YqcA from Escherichia coli
To be Published
|
|
2MOL
 
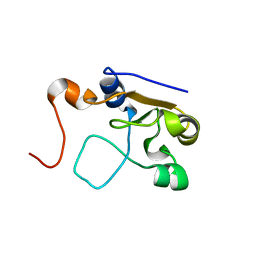 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOM
 
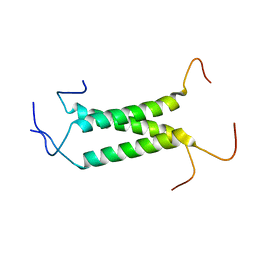 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
2MOP
 
 | | Structure of Bitistatin A | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-04-29 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2MOQ
 
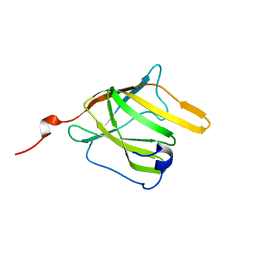 | | Solution Structure and Molecular determinants of Hemoglobin Binding of the first NEAT Domain of IsdB in Staphylococcus aureus | | Descriptor: | Iron-regulated surface determinant protein B | | Authors: | Fonner, B.A, Tripet, B.P, Eilers, B.J, Stanisich, J, Sullivan-Springhetti, R.K, Moore, R, Lui, M, Lei, B, Copie, V. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Molecular Determinants of Hemoglobin Binding of the First NEAT Domain of IsdB in Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
2MOR
 
 | |
2MOT
 
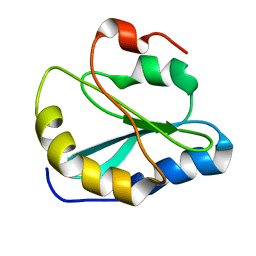 | | Backbone Structure of Actin Depolymerizing Factor (ADF) of Toxoplasma gondii Based on Prot3DNMR Approach | | Descriptor: | Actin depolymerizing factor ADF | | Authors: | Kumar, D, Raikwal, N, Raval, I, Jaiswal, N, Shukla, V, Arora, A. | | Deposit date: | 2014-05-05 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Prot3DNMR: A Simple and Swift Strategy for Backbone Structure Determination of Proteins by NMR
To be Published
|
|
2MOU
 
 | |
2MOV
 
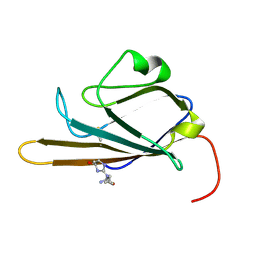 | | Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal Derived AGEs. | | Descriptor: | Advanced glycosylation end product-specific receptor, N~5~-[(5R)-5-methyl-4-oxo-4,5-dihydro-1H-imidazol-2-yl]-L-ornithine | | Authors: | Shekhtman, A, Xue, J, Ray, R, Singer, D, Bohme, D, Burz, D.S, Rai, V, Hoffman, R. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Receptor for Advanced Glycation End Products (RAGE) Specifically Recognizes Methylglyoxal-Derived AGEs.
Biochemistry, 53, 2014
|
|
