2M7O
 
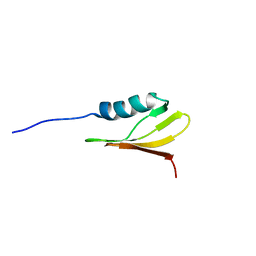 | |
2M7P
 
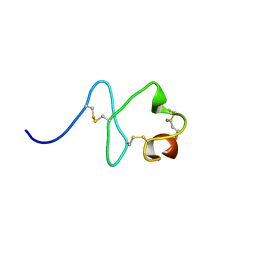 | | RXFP1 utilises hydrophobic moieties on a signalling surface of the LDLa module to mediate receptor activation | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Relaxin receptor 1 | | Authors: | Kong, R.CK, Petrie, E.J, Mohanty, B, Ling, J, Lee, J.C.Y, Gooley, P.R, Bathgate, R.A.D. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.
J.Biol.Chem., 288, 2013
|
|
2M7Q
 
 | | Solution structure of TAX1BP1 UBZ1+2 | | Descriptor: | Tax1-binding protein 1, ZINC ION | | Authors: | Ceregido, M.A, Spinola Amilibia, M, Buts, L, Bravo, J, van Nuland, N.A.J. | | Deposit date: | 2013-04-29 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of TAX1BP1 UBZ1+2 provides insight into target specificity and adaptability.
J.Mol.Biol., 426, 2014
|
|
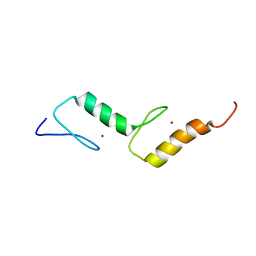
2M7R
 
 | | Nmda receptor antagonist, conantokin bk-b, nmr, 20 structure | | Descriptor: | CONANTOKIN BK-B | | Authors: | Curtice, K.J, Platt, R.J, Skalicky, J.J, Horvath, M.P, Twede, V.D. | | Deposit date: | 2013-04-29 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes.
Toxicon, 81C, 2014
|
|
2M7S
 
 | | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1 | | Descriptor: | Serine/arginine-rich splicing factor 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1
To be Published
|
|
2M7T
 
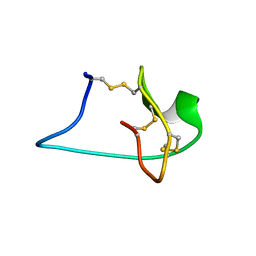 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
2M7U
 
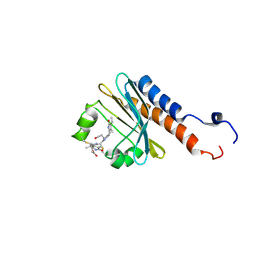 | | Blue Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2M7V
 
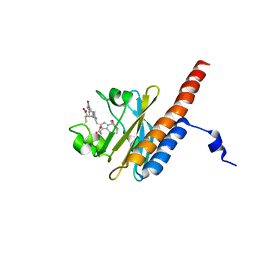 | | Green Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, PHYCOVIOLOBILIN | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2M7W
 
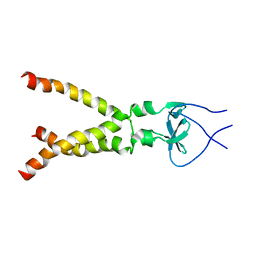 | | Independently verified structure of gp41-M-MAT, a membrane associated MPER trimer from HIV-1 gp41 | | Descriptor: | Envelope glycoprotein | | Authors: | Martin, J.W, Reardon, P.N, Sage, H.S, Moses, D.S, Munir, A.S, Haynes, B.F, Spicer, L.D, Donald, B.R. | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an HIV-1-neutralizing antibody target, the lipid-bound gp41 envelope membrane proximal region trimer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2M7X
 
 | | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2 | | Descriptor: | Na(+)/H(+) antiporter | | Authors: | Ullah, A, Kemp, G, Lee, B, Alves, C, Young, H, Sykes, B.D, Fliegel, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane Segment IV of the Salt Tolerance Protein Sod2.
J.Biol.Chem., 288, 2013
|
|
2M7Y
 
 | | The Mengovirus Leader protein | | Descriptor: | Leader peptide, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Zhao, K.Q, Davis, V, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Full Length Mengovirus Leader
To be Published
|
|
2M7Z
 
 | | Structure of SmTSP2EC2 | | Descriptor: | CD63-like protein Sm-TSP-2 | | Authors: | Mulvenna, J, Jia, X. | | Deposit date: | 2013-05-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure, membrane interactions, and protein binding partners of the tetraspanin Sm-TSP-2, a vaccine antigen from the human blood fluke Schistosoma mansoni
J.Biol.Chem., 289, 2014
|
|
2M80
 
 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
2M83
 
 | |
2M84
 
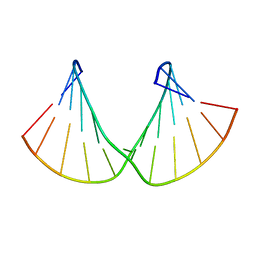 | | Structure of 2'F-RNA/2'F-ANA chimeric duplex | | Descriptor: | 2'F-RNA/2'F-ANA CHIMERIC DUPLEX | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2M85
 
 | | PHD Domain from Human SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, ZINC ION | | Authors: | Machado, L.E.S.F, Pustovalova, Y, Pozhidaeva, A, Almeida, F.C.L, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2013-05-07 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PHD domain from human SHPRH.
J.Biomol.Nmr, 56, 2013
|
|
2M86
 
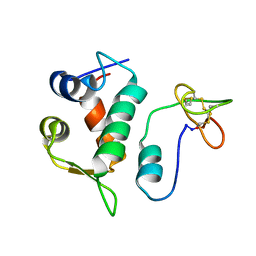 | | Solution structure of Hdm2 with engineered cyclotide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, MCo-PMI | | Authors: | Majumder, S, Ji, Y, Millard, M, Borra, R, Bi, T, Elnagar, A.Y, Neamati, N, Camarero, J.A. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide.
J.Am.Chem.Soc., 135, 2013
|
|
2M87
 
 | |
2M88
 
 | |
2M89
 
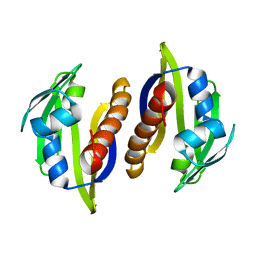 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2M8A
 
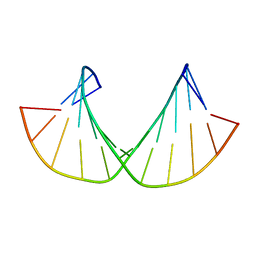 | | 2'F-ANA/2'F-RNA alternated sequences | | Descriptor: | 2'F-RNA/2'F-ANA chimeric duplex | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2M8B
 
 | |
2M8C
 
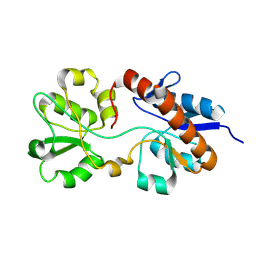 | | The solution NMR structure of E. coli apo-HisJ | | Descriptor: | Cationic amino acid ABC transporter, periplasmic binding protein | | Authors: | Chu, B.C.H, Vogel, H.J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Role of the Two Structural Domains from the Periplasmic Escherichia coli Histidine-binding Protein HisJ.
J.Biol.Chem., 288, 2013
|
|
2M8D
 
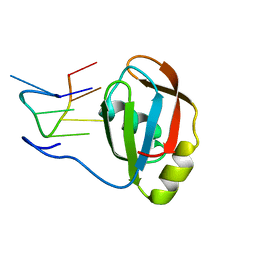 | | Structure of SRSF1 RRM2 in complex with the RNA 5'-UGAAGGAC-3' | | Descriptor: | RNA (5'-R(*UP*GP*AP*AP*GP*GP*AP*C)-3'), Serine/arginine-rich splicing factor 1 | | Authors: | Clery, A, Sinha, R, Anczukow, O, Corrionero, A, Moursy, A, Daubner, G, Valcarcel, J, Krainer, A.R, Allain, F.H.T. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M8E
 
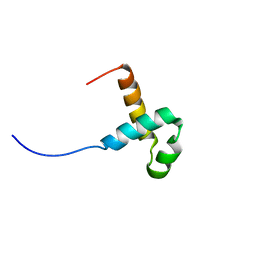 | | NMR structure of the PAI subdomain of Sleeping Beauty transposase | | Descriptor: | SLEEPING BEAUTY TRANSPOSASE | | Authors: | Eubanks, C, Schreifels, J, Aronovich, E, Carlson, D, Hacjkett, P, Nesmelova, I. | | Deposit date: | 2013-05-17 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of Sleeping Beauty transposase binding to DNA.
Protein Sci., 23, 2014
|
|
