7LGL
 
 | |
7LGV
 
 | |
7LHC
 
 | |
7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
7LI2
 
 | | Omega ester peptide pre-fuscimiditide | | Descriptor: | Pre-fuscimiditide peptide | | Authors: | Link, A.J, Elashal, H.E. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Biosynthesis and characterization of fuscimiditide, an aspartimidylated graspetide.
Nat.Chem., 14, 2022
|
|
7LIE
 
 | |
7LIF
 
 | |
7LL7
 
 | | [2]Catenane From MccJ25 Variant G12C G21C | | Descriptor: | GLY-GLY-ALA-GLY-HIS-VAL-PRO-GLU-TYR-PHE, VAL-CYS-ILE-GLY-THR-PRO-ILE-SER-PHE-TYR-CYS | | Authors: | Link, A.J, Schroeder, H.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Dynamic covalent self-assembly of mechanically interlocked molecules solely made from peptides.
Nat.Chem., 13, 2021
|
|
7LNS
 
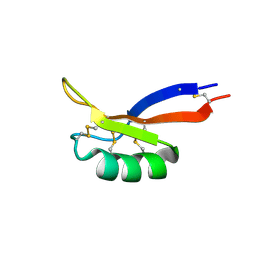 | | NMR solution structure of PsDef2 defensin from P. sylvestris | | Descriptor: | Defensin-2 | | Authors: | Nesmelova, I.V, Kovaleva, V, Hrunyk, N.I, Yusypovych, Y.M, Shalovylo, Y.I. | | Deposit date: | 2021-02-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and function of PsDef2 defensin from Pinus sylvestris.
Structure, 30, 2022
|
|
7LOH
 
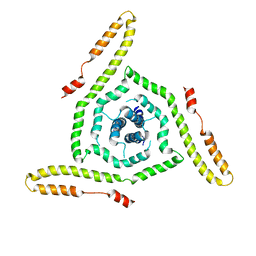 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
7LOI
 
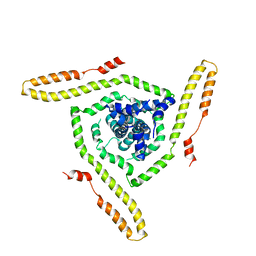 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
7LP4
 
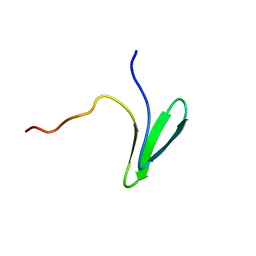 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LQT
 
 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
7LSO
 
 | | L-Phenylseptin | | Descriptor: | L-Phenylseptin peptide | | Authors: | Munhoz, V.H, Verly, R.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of L-Phenylseptin in DPC-d38.
To Be Published
|
|
7LSP
 
 | |
7LU8
 
 | |
7LUW
 
 | |
7LVA
 
 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
7LVE
 
 | |
7LVF
 
 | |
7LVG
 
 | |
7LVN
 
 | |
7LX4
 
 | |
7LXC
 
 | |
