6K5T
 
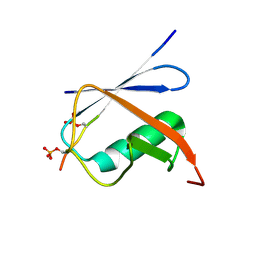 | | Complex of SUMO1 and phosphorylated hcmv protein IE2 | | Descriptor: | 12-mer from Viral transcription factor IE2, Small ubiquitin-related modifier 1 | | Authors: | Tripathi, V, Chatterjee, K.S. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Casein kinase-2-mediated phosphorylation increases the SUMO-dependent activity of the cytomegalovirus transactivator IE2.
J.Biol.Chem., 294, 2019
|
|
6K5W
 
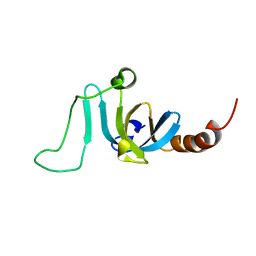 | | Solution structure of the chromodomain of yeast Eaf3 | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eaf3 chromodomain acts as a pH sensor for gene expression by altering its binding affinity for histone methylated-lysine residues.
Biosci.Rep., 40, 2020
|
|
6K7W
 
 | | Solution Structure of the CS1 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
6K84
 
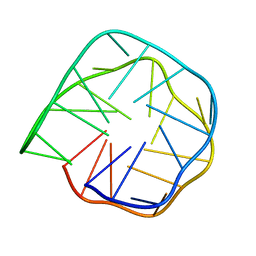 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
6K8Q
 
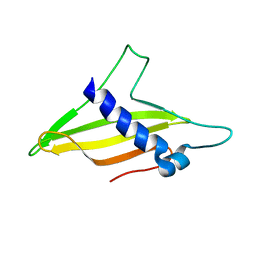 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6KBO
 
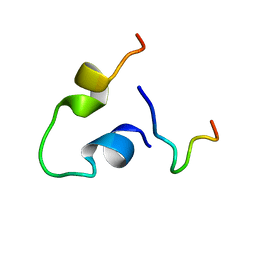 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
6KBV
 
 | |
6KCZ
 
 | |
6KFI
 
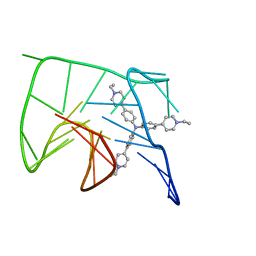 | | NMR solution structure of the 1:1 complex of Tel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA (26-MER) | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KFJ
 
 | | NMR solution structure of the 1:1 complex of wtTel26 G-quadruplex and a tripodal cationic fluorescent probe NBTE | | Descriptor: | 4,4',4''-(nitrilotris(benzene-4,1-diyl))tris(1-ethylpyridin-1-ium) iodide, G-quadruplex DNA wtTel26 | | Authors: | Liu, W, Liu, L.Y, Mao, Z.W. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KH8
 
 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
6KH9
 
 | | Solution structure of bovine insulin amyloid intermediate-1 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Ratha, B.N, Kar, R.K, Brender, J.B, Bhunia, A. | | Deposit date: | 2019-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
6KHA
 
 | | Solution structure of bovine insulin amyloid intermediate-2 | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Ratha, B.N, Kar, R.K, Brender, J.B, Bhunia, A. | | Deposit date: | 2019-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
6KHV
 
 | | Solution Structure of the CS2 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KJO
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the low affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KLM
 
 | | NMR solution structure of Roseltide rT7 | | Descriptor: | Roseltide rT7 | | Authors: | Fan, J.S, Kam, A, Loo, S, Yang, D, Tam, P.J. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation.
J.Biol.Chem., 294, 2019
|
|
6KMY
 
 | | Structure of single disulfide peptide Czon1107-P5A | | Descriptor: | Czon1107-P5A | | Authors: | Sarma, S.P, Madhan Kumar, M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and allosteric activity of a single-disulfide conopeptide fromConus zonatusat human alpha 3 beta 4 and alpha 7 nicotinic acetylcholine receptors.
J.Biol.Chem., 295, 2020
|
|
6KN2
 
 | |
6KN3
 
 | |
6KN4
 
 | |
6KNA
 
 | |
6KNO
 
 | |
