1NHN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NI7
 
 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
1NIL
 
 | | A COMPARISON OF NMR SOLUTION STRUCTURES OF THE RECEPTOR BINDING DOMAINS OF PSEUDOMONAS AERUGINOSA PILI STRAINS PAO, KB7, AND PAK: IMPLICATIONS FOR RECEPTOR BINDING AND SYNTHETIC VACCINE DESIGN | | Descriptor: | PAK PILIN, TRANS | | Authors: | Campbell, A.P, Mcinnes, C, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1995-10-05 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design.
Biochemistry, 34, 1995
|
|
1NIN
 
 | | PLASTOCYANIN FROM ANABAENA VARIABILIS, NMR, 20 STRUCTURES | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Badsberg, U, Jorgensen, A.M.M, Gesmar, H, Led, J.J, Hammerstad-Petersen, J.M, Ulstrup, J. | | Deposit date: | 1996-03-13 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced plastocyanin from the blue-green alga Anabaena variabilis.
Biochemistry, 35, 1996
|
|
1NIX
 
 | |
1NIY
 
 | |
1NIZ
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1NJ0
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | Descriptor: | NPL4, ZINC ION | | Authors: | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
1NK2
 
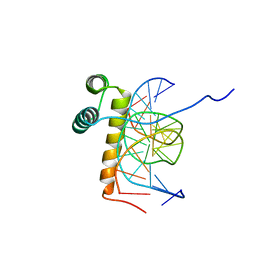 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NKF
 
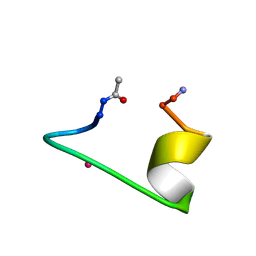 | | CALCIUM-BINDING PEPTIDE, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM-BINDING HEXADECAPEPTIDE, LANTHANUM (III) ION | | Authors: | Sticht, H, Ejchart, A. | | Deposit date: | 1998-03-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix nucleation by a calcium-binding peptide loop.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1NKL
 
 | |
1NKU
 
 | |
1NLA
 
 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
1NM7
 
 | | Solution structure of the ScPex13p SH3 domain | | Descriptor: | Peroxisomal Membrane Protein PAS20 | | Authors: | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
1NMR
 
 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
1NMV
 
 | | Solution structure of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-11 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NMW
 
 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NNV
 
 | |
1NO8
 
 | | SOLUTION STRUCTURE OF THE NUCLEAR FACTOR ALY RBD DOMAIN | | Descriptor: | ALY | | Authors: | Perez-Alvarado, G.C, Martinez-Yamout, M, Allen, M.M, Grosschedl, R, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-01-15 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Nuclear Factor ALY: Insights into Post-Transcriptional
Regulatory and mRNA Nuclear Export Processes
Biochemistry, 42, 2003
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
1NOR
 
 | | TWO-DIMENSIONAL 1H-NMR STUDY OF THE SPATIAL STRUCTURE OF NEUROTOXIN II FROM NAJA OXIANA | | Descriptor: | NEUROTOXIN II | | Authors: | Golovanov, A.P, Utkin, Y.N, Lomize, A.L, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 1993-04-05 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Eur.J.Biochem., 213, 1993
|
|
1NP5
 
 | | (GAC)3 parallel duplex | | Descriptor: | 5'-D(*GP*AP*CP*GP*AP*CP*GP*AP*C)-3' | | Authors: | Zheng, M, Han, X, Gao, X. | | Deposit date: | 2003-01-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Strand polarity of trinucleotide repeat sequences:
NMR studies of parallel/anti-pararell duplex,{d(GAC)3}2
To be Published
|
|
1NP9
 
 | |
1NPQ
 
 | | structure of a rhodamine-labeled N-domain Troponin C mutant (Ca2+ saturated) in complex with skeletal Troponin I 115-131 | | Descriptor: | CALCIUM ION, Troponin C, Troponin I | | Authors: | Mercier, P, Ferguson, R.E, Irving, M, Corrie, J.E.T, Trentham, D.R, Sykes, B.D. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Bifunctional Rhodamine Labeled N-Domain of Troponin C Complexed with the Regulatory
"Switch" Peptide from Troponin I: Implications for in Situ Fluorescence Studies in Muscle Fibers
Biochemistry, 42, 2003
|
|
