5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | Descriptor: | Klebsidin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
5UJ5
 
 | |
5UJG
 
 | | ovGRN12-35_3s | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
5UJH
 
 | | ov-GRN12-34 | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
5UJL
 
 | |
5UJN
 
 | |
5UJQ
 
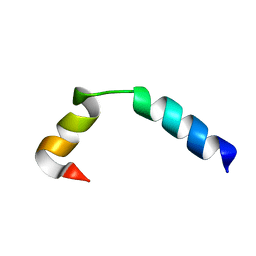 | | NMR Solution Structure of the Two-component Bacteriocin CbnXY | | Descriptor: | Bacteriocin | | Authors: | Acedo, J.Z, Towle, K.M, Lohans, C.T, McKay, R.T, Miskolzie, M, Doerksen, T, Vederas, J.C, Martin-Visscher, L.A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria.
FEBS Lett., 591, 2017
|
|
5UJR
 
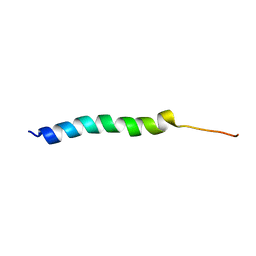 | | NMR Solution Structure of the Two-component Bacteriocin CbnXY | | Descriptor: | Bacteriocin | | Authors: | Acedo, J.Z, Towle, K.M, Lohans, C.T, McKay, R.T, Miskolzie, M, Doerksen, T, Vederas, J.C, Martin-Visscher, L.A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria.
FEBS Lett., 591, 2017
|
|
5UK6
 
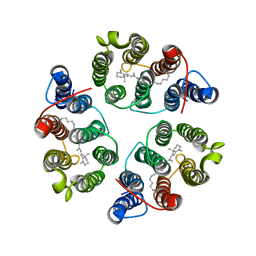 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | Descriptor: | Bacteriorhodopsin | | Authors: | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | Deposit date: | 2017-01-20 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | SOLID-STATE NMR | | Cite: | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
5UKE
 
 | |
5UKZ
 
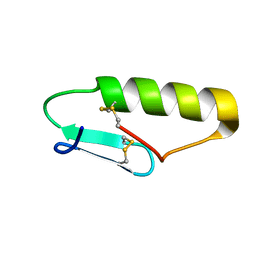 | | NMR Solution structure of chemically synthesized antilisterial Pediocin PA-1 M31L analog. | | Descriptor: | Bacteriocin pediocin PA-1 M31L | | Authors: | Bedard, F, Hammami, R, Zirah, S, Rebuffat, S, Fliss, I, Biron, E. | | Deposit date: | 2017-01-23 | | Release date: | 2018-06-27 | | Method: | SOLUTION NMR | | Cite: | Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof.
Sci Rep, 8, 2018
|
|
5UNK
 
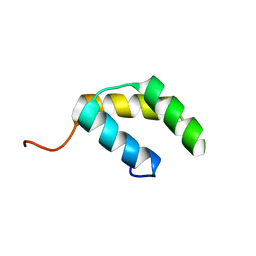 | |
5UOI
 
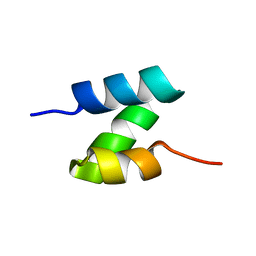 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | Descriptor: | HHH_rd1_0142 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | Descriptor: | EEHEE_rd3_1049 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5URN
 
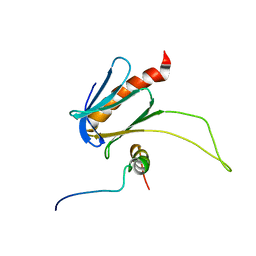 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
5US3
 
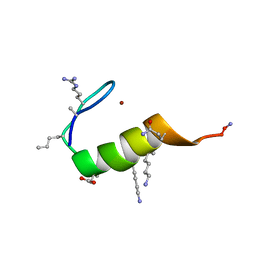 | |
5US5
 
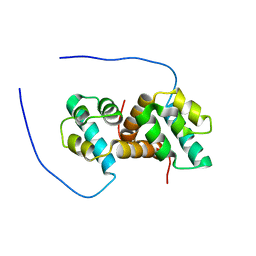 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
5UTG
 
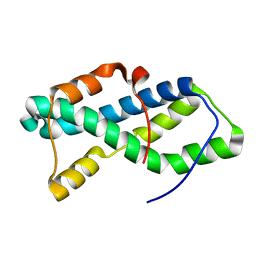 | | Red abalone lysin F104A | | Descriptor: | Egg-lysin | | Authors: | Wilburn, D.B, Tuttle, L.M. | | Deposit date: | 2017-02-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of sperm lysin yields novel insights into molecular dynamics of rapid protein evolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UTV
 
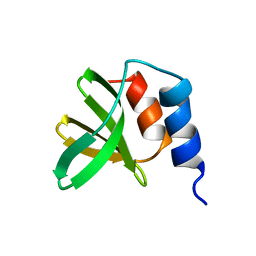 | |
5UY2
 
 | |
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UZ1
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ2
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ3
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
