5TCE
 
 | |
5TCZ
 
 | |
5TGG
 
 | |
5TGW
 
 | | NMR structure of apo-PS1 | | Descriptor: | PS1 | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
5TGY
 
 | | NMR structure of holo-PS1 | | Descriptor: | PS1, [5,10,15,20-tetrakis(trifluoromethyl)porphyrinato(2-)-kappa~4~N~21~,N~22~,N~23~,N~24~]zinc | | Authors: | Polizzi, N.F, Wu, Y. | | Deposit date: | 2016-09-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
5TJ1
 
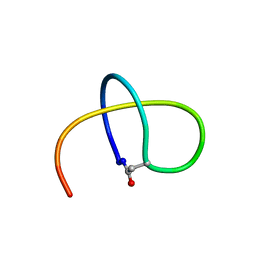 | | Benenodin-1-dC5, state 1 | | Descriptor: | Benenodin-1 | | Authors: | Zong, C, Link, A.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lasso Peptide Benenodin-1 Is a Thermally Actuated [1]Rotaxane Switch.
J. Am. Chem. Soc., 139, 2017
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
5TLR
 
 | | Solution NMR structure of gHwTx-IV | | Descriptor: | Mu-theraphotoxin-Hs2a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Spider peptide toxin HwTx-IV engineered to bind to lipid membranes has an increased inhibitory potency at human voltage-gated sodium channel hNaV1.7.
Biochim. Biophys. Acta, 1859, 2017
|
|
5TM0
 
 | | Solution NMR structures of two alternative conformations of E. coli tryptophan repressor in dynamic equilibrium | | Descriptor: | Trp operon repressor | | Authors: | Harish, B, Swapna, G.V.T, Kornhaber, G.J, Montelione, G.T, Carey, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiple helical conformations of the helix-turn-helix region revealed by NOE-restrained MD simulations of tryptophan aporepressor, TrpR.
Proteins, 85, 2017
|
|
5TMX
 
 | | Solution Structure of SinI, antagonist to the master biofilm-regulator SinR in Bacillus subtilis | | Descriptor: | Protein SinI | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5TN0
 
 | | Solution Structure of the N-terminal DNA-binding domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5TN2
 
 | | Solution Structure of the C-terminal multimerization domain of the master biofilm-regulator SinR from Bacillus subtilis | | Descriptor: | HTH-type transcriptional regulator SinR | | Authors: | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5TP5
 
 | |
5TP6
 
 | |
5TR5
 
 | |
5TRN
 
 | | Solution Structure of a DNA Dodecamer with 8-oxoguanine at the 4th position and 5-methylcytosine at the 9th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*G)-3') | | Authors: | Hoppins, J.J, Gruber, D.R, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5TTB
 
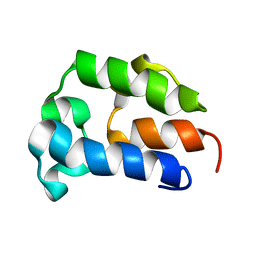 | |
5TTT
 
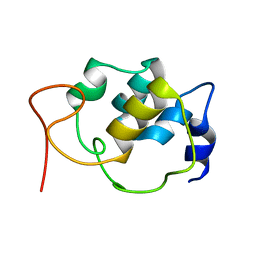 | | Sparse-restraint solution NMR structure of micelle-solubilized cytosolic amino terminal domain of C. elegans mechanosensory ion channel MEC-4 refined by restrained Rosetta | | Descriptor: | Degenerin mec-4 | | Authors: | Everett, J.K, Liu, G, Mao, B, Driscoll, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparse-restraint solution NMR structure of micelle-solubilized
cytosolic amino terminal domain of C. elegans mechanosensory ion channel
MEC-4 refined by restrained Rosetta
To Be Published
|
|
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
5TWI
 
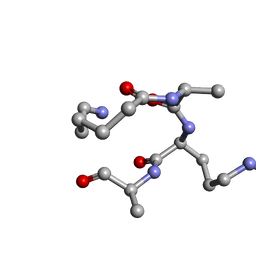 | |
5TWW
 
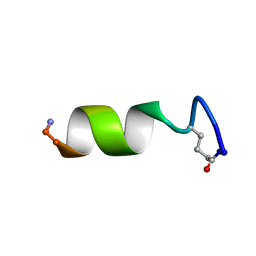 | |
5TX8
 
 | |
5U3H
 
 | |
5U4K
 
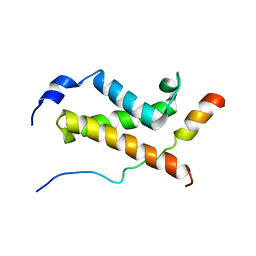 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
5U5S
 
 | |
