2L0C
 
 | | Solution NMR Structure of protein STY4237 (residues 36-120) from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR115 | | Descriptor: | Putative membrane protein | | Authors: | Cort, J.R, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein STY4237 (residues 36-120) from
Salmonella enterica, Northeast Structural Genomics Consortium Target SlR115
To be Published
|
|
2L0D
 
 | | Solution NMR Structure of putative cell surface protein MA_4588 (272-376 domain) from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR254A | | Descriptor: | Cell surface protein | | Authors: | Cort, J.R, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of putative cell surface protein MA_4588
(272-376 domain) from Methanosarcina acetivorans, Northeast Structural
Genomics Consortium Target MvR254A
To be Published
|
|
2L0E
 
 | |
2L0F
 
 | |
2L0G
 
 | |
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2L0J
 
 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
2L0K
 
 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
2L0L
 
 | | DsbB2 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0M
 
 | | DsbB2 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0N
 
 | | DsbB3 peptide structure in 70% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0O
 
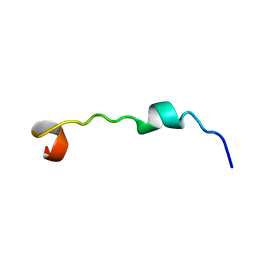 | | DsbB3 peptide structure in 100% TFE | | Descriptor: | Oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | | Authors: | Hwang, S, Hilty, C. | | Deposit date: | 2010-07-08 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding determinants of disulfide bond forming protein B explored by solution nuclear magnetic resonance spectroscopy.
Proteins, 79, 2011
|
|
2L0P
 
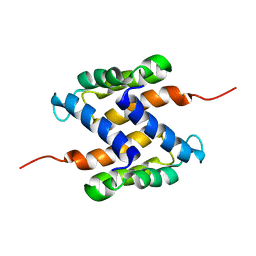 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
2L0Q
 
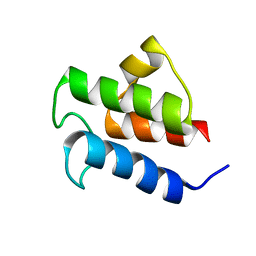 | | NMR Solution Structure of Vibrio harveyi Acyl Carrier Protein (ACP) | | Descriptor: | Acyl carrier protein | | Authors: | Chan, D.I, Chu, B.C.H, Lau, C.K.Y, Hunter, H.N, Byers, D.M, Vogel, H.J. | | Deposit date: | 2010-07-12 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and biophysical characterization of Vibrio harveyi acyl carrier protein A75H: effects of divalent metal ions.
J.Biol.Chem., 285, 2010
|
|
2L0R
 
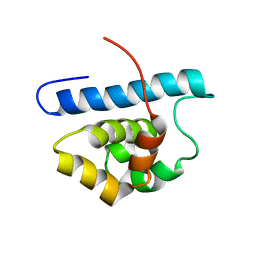 | | Conformational Dynamics of the Anthrax Lethal Factor Catalytic Center | | Descriptor: | Lethal factor | | Authors: | Dalkas, G.A, Chasapis, C.T, Gkazonis, P.V, Bentrop, D.A, Spyroulias, G.A. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational dynamics of the anthrax lethal factor catalytic center.
Biochemistry, 49, 2010
|
|
2L0S
 
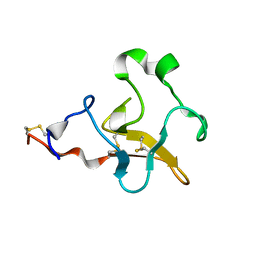 | | Solution Structure of Human Plasminogen Kringle 3 | | Descriptor: | Plasminogen | | Authors: | Christen, M.T, Frank, P, Schaller, J, Llinas, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Human plasminogen kringle 3: solution structure, functional insights, phylogenetic landscape.
Biochemistry, 49, 2010
|
|
2L0T
 
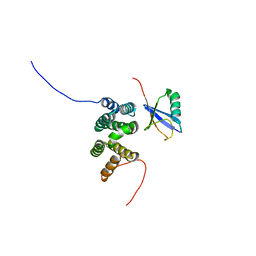 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
2L0W
 
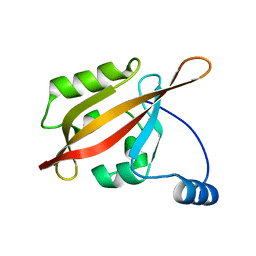 | | Solution NMR structure of the N-terminal PAS domain of HERG potassium channel | | Descriptor: | Potassium voltage-gated channel, subfamily H (Eag-related), member 2, ... | | Authors: | Ng, C.A, Hunter, M.J, Mobli, M, King, G.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2010-07-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Tail of hERG Contains an Amphipathic alpha-Helix That Regulates Channel Deactivation
PLoS ONE, 6, 2011
|
|
2L0X
 
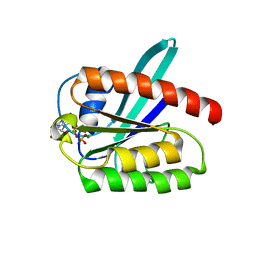 | | Solution structure of the 21 kDa GTPase RHEB bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stoll, R, Heumann, R, Berghaus, C, Kock, G. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ras homolog enriched in brain (Rheb) enhances apoptotic signaling.
J.Biol.Chem., 285, 2010
|
|
2L0Y
 
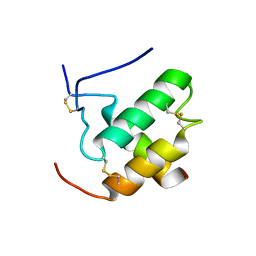 | | Complex hMia40-hCox17 | | Descriptor: | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) pseudogene (COX17), Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Bertini, I, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-11-24 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2L0Z
 
 | |
2L10
 
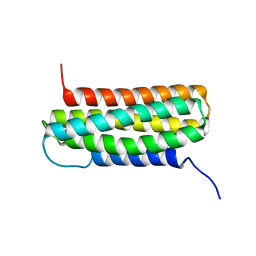 | | Solution Structure of the R6 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Roberts, G.C.K, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.
J.Biol.Chem., 288, 2013
|
|
2L11
 
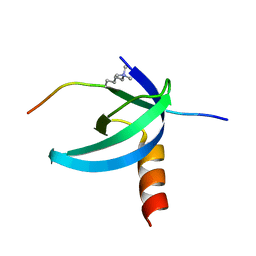 | | Solution NMR structure of the Cbx3 in complex with H3K9me3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L12
 
 | | Solution NMR structure of the chromobox protein 7 with H3K9me3 | | Descriptor: | Chromobox homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Gutmanas, A, Fares, C, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L13
 
 | |
