5YFG
 
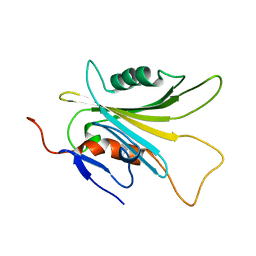 | | SOLUTION STRUCTURE OF HUMAN MOG1 | | Descriptor: | Ran guanine nucleotide release factor | | Authors: | Hu, Q, Liu, Y, Bao, X, Liu, H. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation
J Mol Cell Biol, 10, 2018
|
|
5YHN
 
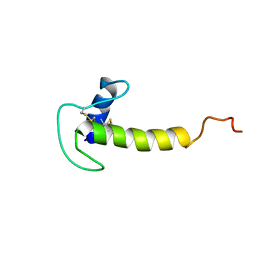 | | Solution structure of the LEKTI Domain 4 | | Descriptor: | cDNA FLJ60407, highly similar to Serine protease inhibitor Kazal-type 5 | | Authors: | Mok, Y.K, Ramesh, K. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Lympho-Epithelial Kazal-type Inhibitor Domains Delay Blood Coagulation by Inhibiting Factor X and XI with Differential Specificity.
Structure, 26, 2018
|
|
5YI4
 
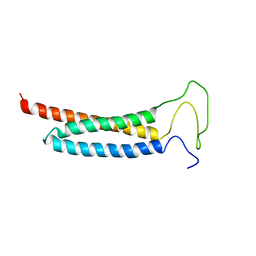 | | Solution Structure of the DISC1/Ndel1 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Nuclear distribution protein nudE-like 1 | | Authors: | Ye, F, Yu, C, Yu, C, Zhang, M. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis.
Neuron, 96, 2017
|
|
5YI9
 
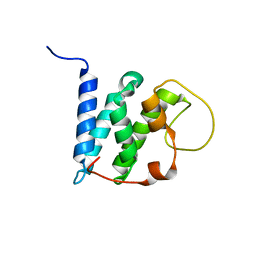 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 56-91) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Cell division cycle-associated 7-like protein | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YIO
 
 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
5YJ4
 
 | |
5YJ5
 
 | |
5YKK
 
 | |
5YKL
 
 | |
5YKQ
 
 | |
5YMY
 
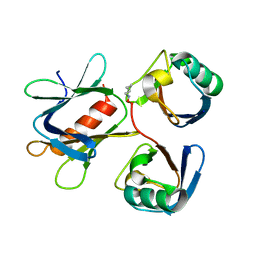 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
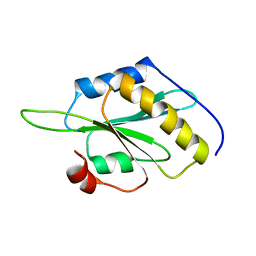
5YNR
 
 | |
5YOZ
 
 | |
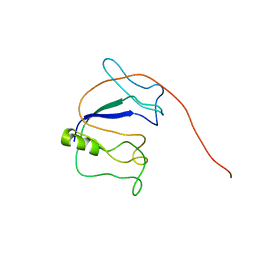
5YQ3
 
 | |
5YXI
 
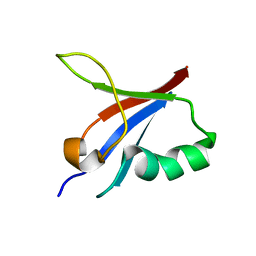 | | Designed protein dRafX6 | | Descriptor: | Designed protein dRafX6 | | Authors: | Liu, R. | | Deposit date: | 2017-12-05 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo sequence redesign of a functional Ras-binding domain globally inverted the surface charge distribution and led to extreme thermostability.
Biotechnol.Bioeng., 118, 2021
|
|
5YZ6
 
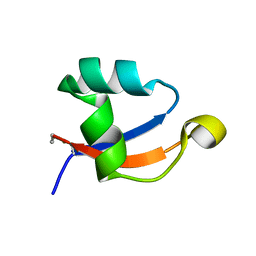 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5YZ9
 
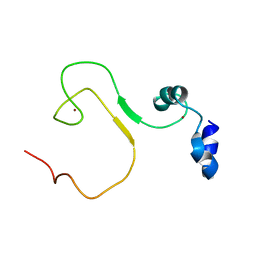 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | Descriptor: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | Authors: | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
5YZK
 
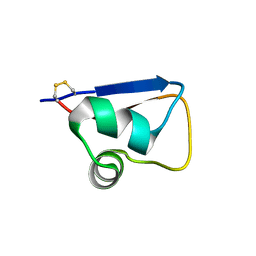 | | Solution structure of LysM domain from a chitinase derived from Volvox carteri | | Descriptor: | Chitinase, lysozyme | | Authors: | Kitaoku, Y, Nishimura, S, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structures and chitin-binding properties of two N-terminal lysin motifs (LysMs) found in a chitinase from Volvox carteri.
Glycobiology, 29, 2019
|
|
5Z1Y
 
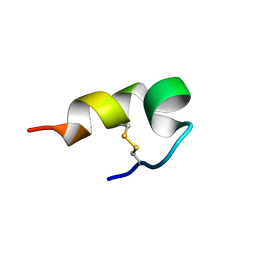 | | mBjAMP1 structure | | Descriptor: | mBjAMP1 peptide | | Authors: | Nam, J.Y, Lee, C.W. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | mBjAMP1 structure
To Be Published
|
|
5Z26
 
 | |
5Z2O
 
 | |
5Z31
 
 | | LPS bound solution structure of WS2-KG18 | | Descriptor: | LYS-ASN-LYS-SER-ARG-VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5Z32
 
 | | LPS bound solution NMR structure of WS2-VR18 | | Descriptor: | VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY-LYS-ASN-LYS-SER-ARG | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
5Z36
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with PDM | | Descriptor: | 11-hydroxy-2-[(2S)-2-hydroxybutan-2-yl]-5-methyl-4H-anthra[1,2-b]pyran-4,7,12-trione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z4B
 
 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
