2LXP
 
 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LXQ
 
 | | Monomeric PilE G-Quadruplex DNA from Neisseria Gonorrhoeae | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*T)-3') | | Authors: | Kuryavyi, V.V, Patel, D.J. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RecA-Binding pilE G4 Sequence Essential for Pilin Antigenic Variation Forms Monomeric and 5' End-Stacked Dimeric Parallel G-Quadruplexes.
Structure, 20, 2012
|
|
2LXR
 
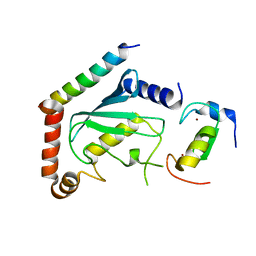
 | | Solution structure of HP1264 from Helicobacter pylori | | Descriptor: | NADH dehydrogenase I subunit E | | Authors: | Lee, K. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HP1264, Complex I subunit E from Helicobacter pylori, reveals a novel fold for FMN binding protein
To be Published
|
|
2LXS
 
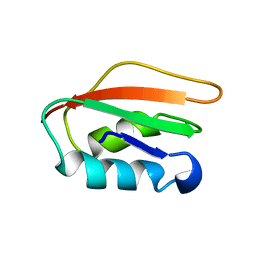
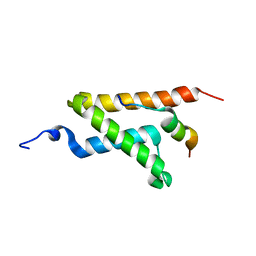 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2LXT
 
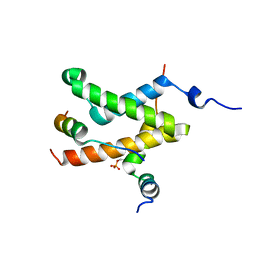 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2LXU
 
 | | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Ramelot, T.A, Yang, Y, Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A
To be Published
|
|
2LXV
 
 | |
2LXW
 
 | |
2LXX
 
 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 2, isoform c | | Authors: | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
2LXY
 
 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
2LXZ
 
 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | Descriptor: | Defensin-5 | | Authors: | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
2LY0
 
 | | Solution NMR structure of the influenza A virus S31N mutant (19-49) in presence of drug M2WJ332 | | Descriptor: | (3S,5S,7S)-N-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methyl}tricyclo[3.3.1.1~3,7~]decan-1-aminium, Membrane ion channel M2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LY1
 
 | |
2LY2
 
 | |
2LY3
 
 | |
2LY4
 
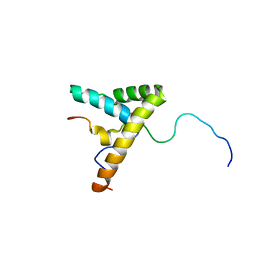 | | HMGB1-facilitated p53 DNA binding occurs via HMG-box/p53 transactivation domain interaction and is regulated by the acidic tail | | Descriptor: | Cellular tumor antigen p53, High mobility group protein B1 | | Authors: | Rowell, J.P, Simpson, K.L, Stott, K, Watson, M, Thomas, J.O. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | HMGB1-Facilitated p53 DNA Binding Occurs via HMG-Box/p53 Transactivation Domain Interaction, Regulated by the Acidic Tail.
Structure, 20, 2012
|
|
2LY5
 
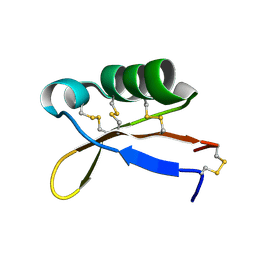 | | Refined solution structure of recombinant brazzein | | Descriptor: | Defensin-like protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Tonelli, M, Markley, J.L, Assadi-Porter, F.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein.
Proteins, 81, 2013
|
|
2LY6
 
 | | Refined solution structure of recombinant brazzein at low temperature | | Descriptor: | Defensin-like protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Tonelli, M, Markley, J.L, Assadi-Porter, F.M. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein.
Proteins, 81, 2013
|
|
2LY7
 
 | | B-flap domain of RNA polymerase (B. subtilis) | | Descriptor: | DNA-directed RNA polymerase subunit beta | | Authors: | Mobli, M. | | Deposit date: | 2012-09-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase-induced remodelling of NusA produces a pause enhancement complex.
Nucleic Acids Res., 43, 2015
|
|
2LY8
 
 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
2LY9
 
 | | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F | | Descriptor: | Zinc fingers and homeoboxes protein 1 | | Authors: | Xu, X, Eletsky, A, Mills, J.L, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F
To be Published
|
|
2LYA
 
 | |
2LYB
 
 | |
2LYC
 
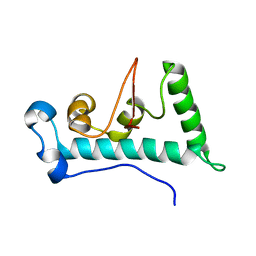 | | Structure of C-terminal domain of Ska1 | | Descriptor: | Spindle and kinetochore-associated protein 1 homolog | | Authors: | Boeszoermenyi, A, Schmidt, J.C, Markus, M, Oberer, M, Cheeseman, I.M, Wagner, G, Arthanari, H. | | Deposit date: | 2012-09-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments.
Dev.Cell, 23, 2012
|
|
2LYD
 
 | |
