2LMV
 
 | | Androcam at high calcium with three explicit Ca2+ | | Descriptor: | CALCIUM ION, Calmodulin-related protein 97A | | Authors: | Joshi, M.K, Moran, S.T, Beckingham, K.M, Mackenzie, K.R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of androcam supports specialized interactions with myosin VI.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LMZ
 
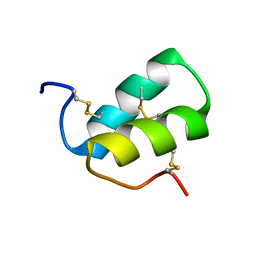 | |
2LN0
 
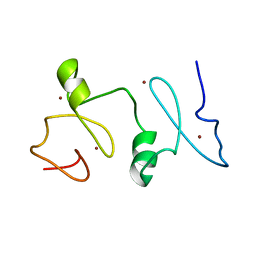 | | Structure of MOZ | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Qiu, Y. | | Deposit date: | 2011-12-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription.
Genes Dev., 26, 2012
|
|
2LN3
 
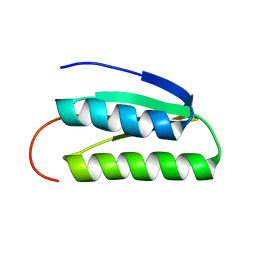 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, IF3-like fold, Northeast Structural Genomics Consortium Target OR135 (CASD target) | | Descriptor: | DE NOVO DESIGNED PROTEIN OR135 | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LN4
 
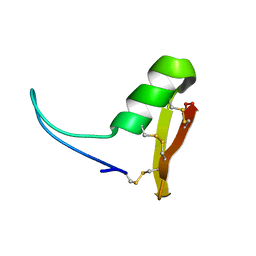 | |
2LN7
 
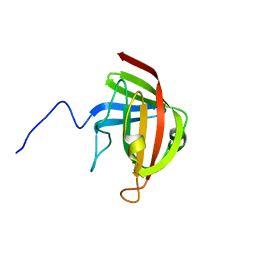 | |
2LN8
 
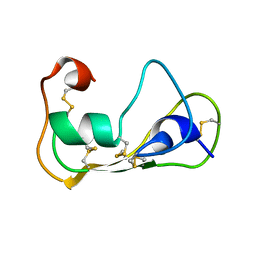 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
2LNA
 
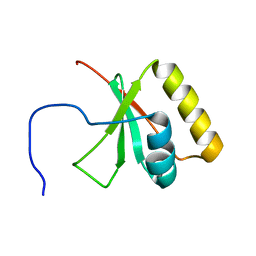 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2LNB
 
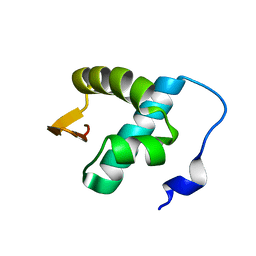 | | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A. | | Descriptor: | Z-DNA-binding protein 1 | | Authors: | Yang, Y, Ramelot, T.A, Hamilton, K, Kohan, E, Wang, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A
To be Published
|
|
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
2LND
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, PFK fold, Northeast Structural Genomics Consortium Target OR134 | | Descriptor: | DE NOVO DESIGNED PROTEIN, PFK fold | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR134
To be Published
|
|
2LNE
 
 | |
2LNF
 
 | |
2LNG
 
 | |
2LNH
 
 | | Enterohaemorrhagic E. coli (EHEC) exploits a tryptophan switch to hijack host F-actin assembly | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, Neural Wiskott-Aldrich syndrome protein, Secreted effector protein EspF(U) | | Authors: | Aitio, O, Hellman, M, Skehan, B, Kesti, T, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2011-12-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Structure, 20, 2012
|
|
2LNI
 
 | | Solution NMR Structure of Stress-induced-phosphoprotein 1 STI1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4403E | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Tang, Y, Liu, G, Hamilton, K, Ciccosanti, C, Shastry, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4403E
To be Published
|
|
2LNJ
 
 | |
2LNK
 
 | |
2LNL
 
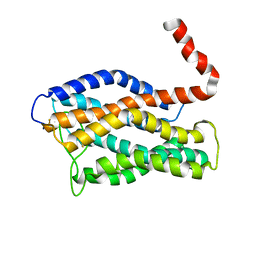 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
2LNM
 
 | |
2LNQ
 
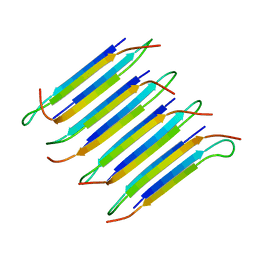 | | 40-residue D23N beta amyloid fibril | | Descriptor: | P3(40) | | Authors: | Qiang, W, Yau, W, Luo, Y, Mattson, M.P, Tycko, R. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Antiparallel beta-sheet architecture in Iowa-mutant beta-amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LNS
 
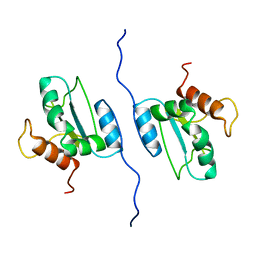 | | Solution structure of AGR2 residues 41-175 | | Descriptor: | Anterior gradient protein 2 homolog | | Authors: | Patel, P, Clarke, C.J, Barraclough, D.L, Rudland, P.S, Barraclough, R, Lian, L. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metastasis-promoting anterior gradient 2 protein has a dimeric thioredoxin fold structure and a role in cell adhesion.
J.Mol.Biol., 425, 2013
|
|
2LNT
 
 | | Solution structure of E60A mutant AGR2 | | Descriptor: | Anterior gradient protein 2 homolog | | Authors: | Patel, P, Clarke, C.J, Barraclough, D.L, Rudland, P.S, Barraclough, R, Lian, L. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metastasis-promoting anterior gradient 2 protein has a dimeric thioredoxin fold structure and a role in cell adhesion.
J.Mol.Biol., 425, 2013
|
|
2LNU
 
 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LNV
 
 | |
