2EVN
 
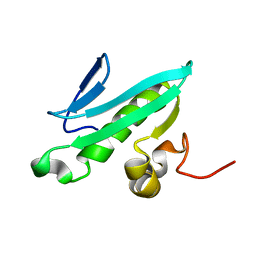 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
2EVQ
 
 | |
2EVY
 
 | | GNYA tetranucleotide loops found in poliovirus oriL by in vivo SELEX (un)expectedly form a YNMG-like structure | | Descriptor: | Poliovirus 5'NTR cloverleaf stem loop D mutant | | Authors: | Melchers, W.J.G, Zoll, J, Tessari, M, Bakhmutov, D.V, Gmyl, A.P, Agol, V.I, Heus, H.A. | | Deposit date: | 2005-11-01 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A GCUA tetranucleotide loop found in the poliovirus oriL by in vivo SELEX (un)expectedly forms a YNMG-like structure: Extending the YNMG family with GYYA.
RNA, 12, 2006
|
|
2EVZ
 
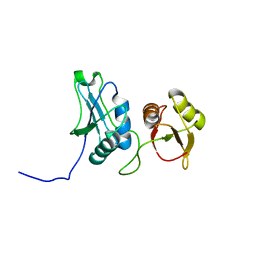 | |
2EW3
 
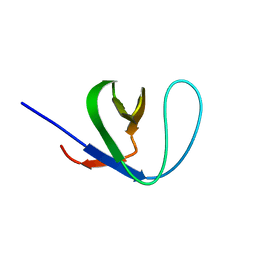 | |
2EW4
 
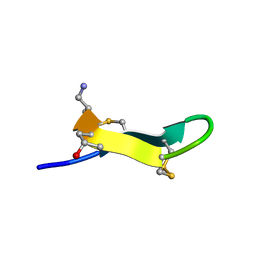 | | Solution structure of MrIA | | Descriptor: | MrIA | | Authors: | Nilsson, K.P, Lovelace, E.S, Caesar, C.E, Tynngard, N, Alewood, P.F, Johansson, H.M, Sharpe, I.A, Lewis, R.J, Daly, N.L, Craik, D.J. | | Deposit date: | 2005-11-02 | | Release date: | 2005-11-15 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution structure of chi-conopeptide MrIA, a modulator of the human norepinephrine transporter
Biopolymers, 80, 2005
|
|
2EW9
 
 | |
2EWL
 
 | |
2EXD
 
 | | The solution structure of the C-terminal domain of a nfeD homolog from Pyrococcus horikoshii | | Descriptor: | nfeD short homolog | | Authors: | Kuwahara, Y, Ohno, A, Morii, T, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold.
Protein Sci., 17, 2008
|
|
2EXF
 
 | |
2EXG
 
 | | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain | | Descriptor: | (5R)-2-SULFANYL-5-[4-(TRIFLUOROMETHYL)BENZYL]-1,3-THIAZOL-4-ONE, Afadin | | Authors: | Joshi, M, Vargas, C, Boisguerin, P, Krause, G, Schade, M, Oschkinat, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2EXN
 
 | | Solution structure for the protein coded by gene locus BB0938 of Bordetella bronchiseptica. Northeast Structural Genomics target BoR11. | | Descriptor: | Hypothetical protein BoR11 | | Authors: | Rossi, P, Ramelot, T, Xiao, R, Ho, C.K, Ma, L.-C, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-08 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C, and (15)N Resonance Assignments for the Protein Coded by Gene Locus BB0938 of Bordetella bronchiseptica
J.Biomol.NMR, 33, 2005
|
|
2EYV
 
 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYW
 
 | |
2EYX
 
 | |
2EYY
 
 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYZ
 
 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EZ5
 
 | | Solution Structure of the dNedd4 WW3* Domain- Comm LPSY Peptide Complex | | Descriptor: | Commissureless LPSY Peptide, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Kanelis, V, Bruce, M.C, Skrynnikov, N.R, Rotin, D, Forman-Kay, J.D. | | Deposit date: | 2005-11-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for High-Affinity Binding in a Nedd4 WW3(*) Domain-Comm PY Motif Complex
Structure, 14, 2006
|
|
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZL
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZM
 
 | |
2EZO
 
 | |
2EZW
 
 | |
