1CEJ
 
 | | SOLUTION STRUCTURE OF AN EGF MODULE PAIR FROM THE PLASMODIUM FALCIPARUM MEROZOITE SURFACE PROTEIN 1 | | Descriptor: | PROTEIN (MEROZOITE SURFACE PROTEIN 1) | | Authors: | Morgan, W.D, Birdsall, B, Frenkiel, T.A, Gradwell, M.G, Burghaus, P.A, Syed, S.E.H, Uthaipibull, C, Holder, A.A, Feeney, J. | | Deposit date: | 1999-03-08 | | Release date: | 1999-05-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1.
J.Mol.Biol., 289, 1999
|
|
1CEY
 
 | | ASSIGNMENTS, SECONDARY STRUCTURE, GLOBAL FOLD, AND DYNAMICS OF CHEMOTAXIS Y PROTEIN USING THREE-AND FOUR-DIMENSIONAL HETERONUCLEAR (13C,15N) NMR SPECTROSCOPY | | Descriptor: | CHEY | | Authors: | Moy, F.J, Lowry, D.F, Matsumura, P, Dahlquist, F.W, Krywko, J.E, Domaille, P.J. | | Deposit date: | 1994-11-23 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CFA
 
 | | SOLUTION STRUCTURE OF A SEMI-SYNTHETIC C5A RECEPTOR ANTAGONIST AT PH 5.2, 303K, NMR, 20 STRUCTURES | | Descriptor: | COMPLEMENT 5A SEMI-SYNTHETIC ANTAGONIST, SYNTHETIC N-TERMINAL TAIL | | Authors: | Zhang, X, Boyar, W, Galakatos, N, Gonnella, N.C. | | Deposit date: | 1996-09-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding.
Protein Sci., 6, 1997
|
|
1CFC
 
 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFE
 
 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
1CFG
 
 | |
1CFL
 
 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(64T)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-03-19 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer duplex containing the stable 3' T.G base pair of the pyrimidine(6-4)pyrimidone photoproduct [(6-4) adduct]: implications for the highly specific 3' T --> C transition of the (6-4) adduct.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CFP
 
 | |
1CHL
 
 | | NMR SEQUENTIAL ASSIGNMENTS AND SOLUTION STRUCTURE OF CHLOROTOXIN, A SMALL SCORPION TOXIN THAT BLOCKS CHLORIDE CHANNELS | | Descriptor: | CHLOROTOXIN | | Authors: | Lippens, G, Najib, J, Wodak, S.J, Tartar, A. | | Deposit date: | 1994-11-09 | | Release date: | 1995-02-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels.
Biochemistry, 34, 1995
|
|
1CHV
 
 | |
1CIR
 
 | | COMPLEX OF TWO FRAGMENTS OF CI2 [(1-40)(DOT)(41-64)] | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Davis, B.J, Fersht, A.R. | | Deposit date: | 1995-10-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards the complete structural characterization of a protein folding pathway: the structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism
Structure Fold.Des., 1, 1996
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1CK2
 
 | |
1CKK
 
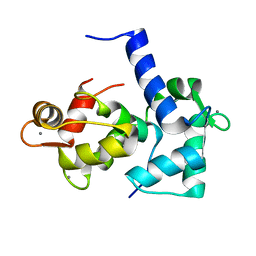 | | CALMODULIN/RAT CA2+/CALMODULIN DEPENDENT PROTEIN KINASE FRAGMENT | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase kinase 1, Calmodulin-1 | | Authors: | Osawa, M, Tokumitsu, H, Swindells, M.B, Kurihara, H, Orita, M, Shibanuma, T, Furuya, T, Ikura, M. | | Deposit date: | 1998-11-20 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase.
Nat.Struct.Biol., 6, 1999
|
|
1CKR
 
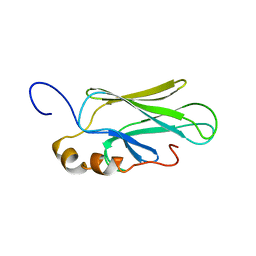 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
1CKV
 
 | |
1CKW
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKX
 
 | | Cystic fibrosis transmembrane conductance regulator: Solution structures of peptides based on the Phe508 region, the most common site of disease-causing Delta-F508 mutation | | Descriptor: | Cystic fibrosis transmembrane conductance regulator (CFTR) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKY
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKZ
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CL4
 
 | |
1CLB
 
 | |
1CLD
 
 | | DNA-binding protein | | Descriptor: | CADMIUM ION, CD2-LAC9 | | Authors: | Gardner, K.H, Coleman, J.E. | | Deposit date: | 1995-06-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain.
Nat.Struct.Biol., 2, 1995
|
|
1CLH
 
 | |
1CMF
 
 | | NMR SOLUTION STRUCTURE OF APO CALMODULIN CARBOXY-TERMINAL DOMAIN | | Descriptor: | CALMODULIN (VERTEBRATE) | | Authors: | Finn, B.E, Evenas, J, Drakenberg, T, Waltho, J.P, Thulin, E, Forsen, S. | | Deposit date: | 1995-07-19 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural changes and domain autonomy in calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
