1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UPH
 
 | | HIV-1 Myristoylated Matrix | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Loeliger, E, Luncsford, P, Kinde, I, Beckett, D, Summers, M.F. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-08 | | Last modified: | 2025-04-09 | | Method: | SOLUTION NMR | | Cite: | Entropic Switch Regulates Myristate Exposure in the HIV-1 Matrix Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UQA
 
 | |
1UQB
 
 | |
1UQC
 
 | |
1UQD
 
 | |
1UQE
 
 | |
1UQF
 
 | |
1UQG
 
 | |
1UQV
 
 | | SAM domain from Ste50p | | Descriptor: | STE50 PROTEIN | | Authors: | Grimshaw, S.J, Mott, H.R, Stott, K.M, Nielsen, P.R, Evetts, K.A, Hopkins, L.J, Nietlispach, D, Owen, D. | | Deposit date: | 2003-10-20 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sterile {Alpha} Motif (Sam) Domain of the Saccharomyces Cerevisiae Mitogen-Activated Protein Kinase Pathway-Modulating Protein Ste50 and Analysis of its Interaction with the Ste11 Sam
J.Biol.Chem., 279, 2004
|
|
1URE
 
 | |
1UTR
 
 | | UTEROGLOBIN-PCB COMPLEX (REDUCED FORM) | | Descriptor: | 4,4'-BIS([H]METHYLSULFONYL)-2,2',5,5'-TETRACHLOROBIPHENYL, UTEROGLOBIN | | Authors: | Hard, T, Barnes, H.J, Larsson, C, Gustafsson, J.-A, Lund, J. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mammalian PCB-binding protein in complex with a PCB.
Nat.Struct.Biol., 2, 1995
|
|
1UTS
 
 | | Designed HIV-1 TAR Binding Ligand | | Descriptor: | N-[2-(3-AMINOPROPOXY)-5-(1H-INDOL-5-YL)BENZYL]-N-(2-PIPERAZIN-1-YLETHYL)AMINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Murchie, A.I.H, Aboul-Ela, F, Karn, J. | | Deposit date: | 2003-12-10 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA.
J.Mol.Biol., 336, 2004
|
|
1UUC
 
 | |
1UUD
 
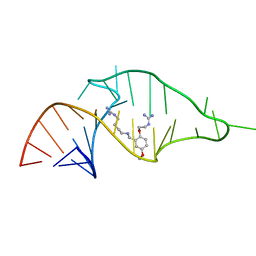 | | NMR structure of a synthetic small molecule, rbt203, bound to HIV-1 TAR RNA | | Descriptor: | N-[2-(2-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-4-METHOXYPHENOXY)ETHYL]GUANIDINE, RNA (5'-(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP *CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C) -3') | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
1UUI
 
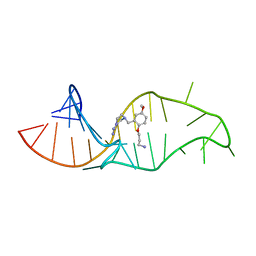 | | NMR structure of a synthetic small molecule, rbt158, bound to HIV-1 TAR RNA | | Descriptor: | 4-[AMINO(IMINO)METHYL]-1-[2-(3-AMMONIOPROPOXY)-5-METHOXYBENZYL]PIPERAZIN-1-IUM, 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*AP*GP*CP* CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*UP*CP*UP*GP*CP*C)-3' | | Authors: | Davis, B, Afshar, M, Varani, G, Karn, J, Murchie, A.I.H, Lentzen, G, Drysdale, M.J, Potter, A.J, Bower, J, Aboul-Ela, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Inhibitors of HIV-1 Tar RNA Through the Stabilisation of Electrostatic "Hot Spots"
J.Mol.Biol., 336, 2004
|
|
1UUU
 
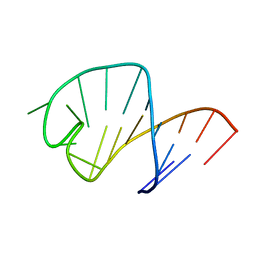 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1UW0
 
 | |
1UWD
 
 | | NMR STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM THERMOTOGA MARITIMA (TM0487), WHICH BELONGS TO THE DUF59 FAMILY. | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
1UWO
 
 | |
1UXC
 
 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1UYA
 
 | | THE SOLUTION STRUCTURE OF THE A-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER A | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1UYB
 
 | | THE SOLUTION STRUCTURE OF THE B-FORM OF UROGUANYLIN-16 NMR, 10 STRUCTURES | | Descriptor: | UROGUANYLIN-16, ISOMER B | | Authors: | Marx, U.C, Adermann, K, Forssmann, W.-G, Roesch, P. | | Deposit date: | 1997-09-11 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | One peptide, two topologies: structure and interconversion dynamics of human uroguanylin isomers.
J.Pept.Res., 52, 1998
|
|
1UZC
 
 | | THE STRUCTURE OF AN FF DOMAIN FROM HUMAN HYPA/FBP11 | | Descriptor: | HYPOTHETICAL PROTEIN FLJ21157 | | Authors: | Allen, M.D, Jemth, P, Friedler, A, Schon, O, Bycroft, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of an Ff Domain from Human Hypa/Fbp11.
J.Mol.Biol., 323, 2002
|
|
1V1C
 
 | |
