5KGQ
 
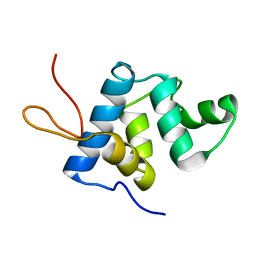 | | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | D'Andrea, E.D, Retel, J.S, Diehl, A, Schmieder, P, Oschkinat, H, Pires, J.R. | | Deposit date: | 2016-06-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi.
J.Struct.Biol., 213, 2021
|
|
5KGV
 
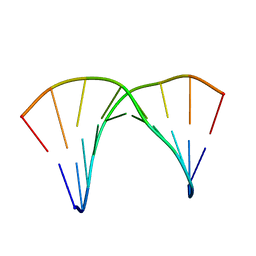 | |
5KGY
 
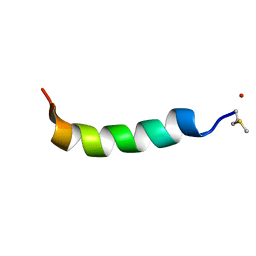 | | Phenol-soluble modulin Alpha 3 | | Descriptor: | Phenol-soluble modulin alpha 3 peptide | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5KGZ
 
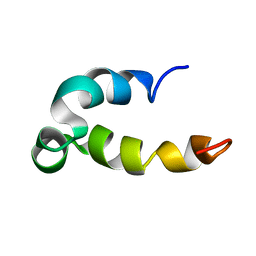 | | Phenol-soluble modulin Beta2 | | Descriptor: | Modulin Beta2 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5KH8
 
 | |
5KHB
 
 | | Structure of Phenol-soluble modulin Alpha1 | | Descriptor: | PSM Alpha1 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Miskolzie, M, van Belkum, M.J, Vederas, J.C. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5KI0
 
 | | NMR structure of human antimicrobial peptide KAMP-19 | | Descriptor: | Antimicrobial peptide KAMP-19 | | Authors: | Wang, G. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane-Active Epithelial Keratin 6A Fragments (KAMPs) Are Unique Human Antimicrobial Peptides with a Non-alpha beta Structure.
Front Microbiol, 7, 2016
|
|
5KI4
 
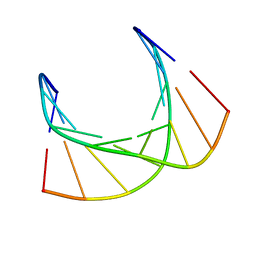 | |
5KI5
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KI7
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIB
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIE
 
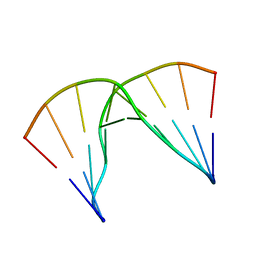 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*GP*GP*A)-R(P*G)-D(P*CP*TP*C)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIF
 
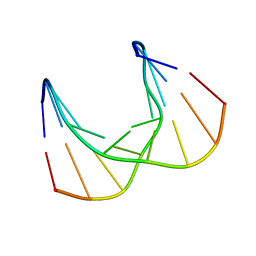 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIH
 
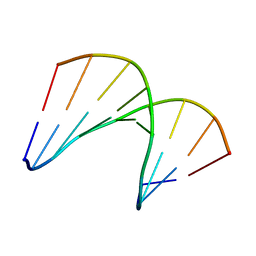 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIZ
 
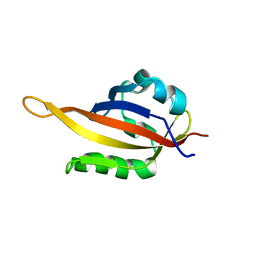 | |
5KJ3
 
 | | Connexin 26 WT peptide NMR Structure | | Descriptor: | Gap junction beta-2 protein | | Authors: | Dowd, T.L, Bargiello, T.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
5KJG
 
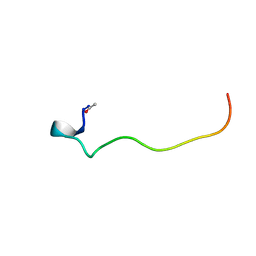 | | Connexin 26 G12R mutant NMR structure | | Descriptor: | Gap junction beta-2 protein | | Authors: | Dowd, T.L, Bargiello, T.A. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
5KK3
 
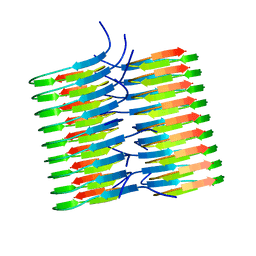 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
5KK9
 
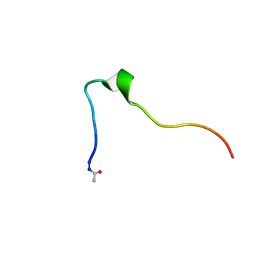 | | Connexin 32 G12R N-Terminal Mutant, | | Descriptor: | Gap junction beta-1 protein | | Authors: | Dowd, T.L, Barigello, T.A. | | Deposit date: | 2016-06-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
5KKM
 
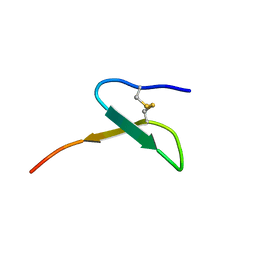 | | Con-Vc11-22 | | Descriptor: | O2_contryphan_Vc1 prepropeptide | | Authors: | Chittoor, B, Krishnarjuna, B, MacRaild, C.A, Robinson, S.D. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Single Disulfide-Directed beta-Hairpin Fold. Dynamics, Stability, and Engineering.
Biochemistry, 56, 2017
|
|
5KMZ
 
 | |
5KNW
 
 | |
5KP0
 
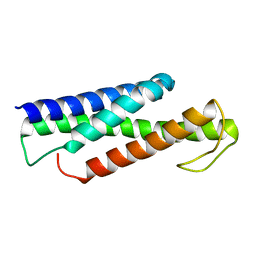 | | Recognition and targeting mechanisms by chaperones in flagella assembly and operation | | Descriptor: | Flagellar protein FliT,Flagellum-specific ATP synthase | | Authors: | Khanra, N.K, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition and targeting mechanisms by chaperones in flagellum assembly and operation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KPE
 
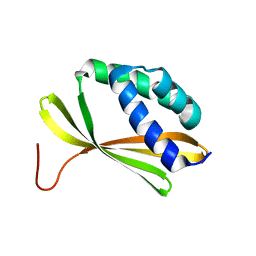 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KPH
 
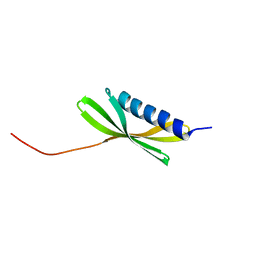 | |
