2KDW
 
 | |
2KDX
 
 | | Solution structure of HypA protein | | Descriptor: | Hydrogenase/urease nickel incorporation protein hypA, ZINC ION | | Authors: | Xia, W, Li, H, Sze, K.-H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites
J.Am.Chem.Soc., 131, 2009
|
|
2KDY
 
 | | NMR structure of LP2086-B01 | | Descriptor: | Factor H binding protein variant B01_001 | | Authors: | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
2KDZ
 
 | | Structure of the R2R3 DNA binding domain of MYB1 protein from protozoan parasite trichomonas vaginalis in complex with MRE-1/MRE-2R DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*TP*A)-3', 5'-D(*TP*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*TP*CP*TP*T)-3', MYB24 | | Authors: | Lou, Y.C, Wei, S.Y, Rajasekaran, M, Chou, C.C, Hsu, H.M, Tai, J.H, Chen, C. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 2009
|
|
2KE0
 
 | | Solution structure of peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Zheng, S, Leeper, T, Napuli, A, Nakazawa, S.H, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of a Burkholderia pseudomallei immunophilin-inhibitor complex reveals new approaches to antimicrobial development.
Biochem.J., 437, 2011
|
|
2KE1
 
 | | Molecular Basis of non-modified histone H3 tail Recognition by the First PHD Finger of Autoimmune Regulator | | Descriptor: | Autoimmune regulator, H3K4me0, ZINC ION | | Authors: | Chignola, F, Gaetani, M, Rebane, A, Org, T, Mollica, L, Zucchelli, C, Spitaleri, A, Mannella, V, Peterson, P, Musco, G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation
Nucleic Acids Res., 37, 2009
|
|
2KE3
 
 | | PC1/3 DCSG sorting domain in CHAPS | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KE4
 
 | |
2KE5
 
 | | Solution structure and dynamics of the small GTPase Ralb in its active conformation: significance for effector protein binding | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-B | | Authors: | Fenwick, R, Prasannan, S, Campbell, L.J, Nietlispach, D, Evetts, K.A, Camonis, J, Mott, H.R, Owen, D. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding
Biochemistry, 48, 2009
|
|
2KE6
 
 | |
2KE7
 
 | | NMR structure of the first SAM domain from AIDA1 | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B | | Authors: | Donaldson, L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the first SAM domain from AIDA1
To be Published
|
|
2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
2KE9
 
 | |
2KEB
 
 | | NMR solution structure of the N-terminal domain of the DNA polymerase alpha p68 subunit | | Descriptor: | DNA polymerase subunit alpha B | | Authors: | Huang, H, Weiner, B.E, Zhang, H, Fuller, B.E, Gao, Y, Wile, B.M, Chazin, W.J, Fanning, E. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.
J.Biol.Chem., 285, 2010
|
|
2KEC
 
 | |
2KED
 
 | |
2KEE
 
 | |
2KEF
 
 | | Solution NMR structures of human hepcidin at 325K | | Descriptor: | Hepcidin | | Authors: | Jordan, J.B, Poppe, L, Hainu, M, Arvedson, T, Syed, R, Li, V, Kohno, H, Kim, H, Miranda, L.P, Cheetham, J, Sasu, B.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2KEG
 
 | | NMR structure of Plantaricin K in DPC-micelles | | Descriptor: | PlnK | | Authors: | Rogne, P, Haugen, M, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two-peptide bacteriocin plantaricin JK.
Peptides, 30, 2009
|
|
2KEH
 
 | |
2KEI
 
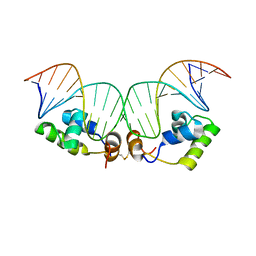 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEJ
 
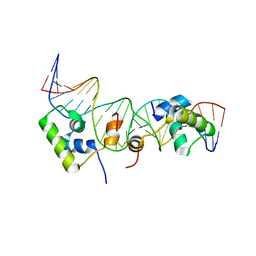 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEK
 
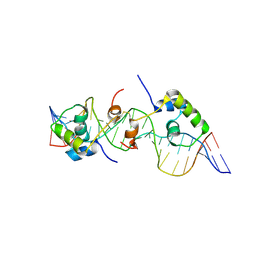 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEL
 
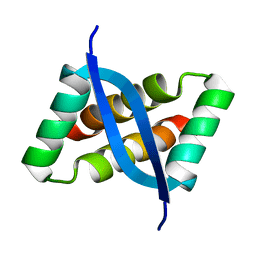 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
2KEM
 
 | | Extended structure of citidine deaminase domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Harjes, E, Gross, P.J, Chen, K, Lu, Y, Shindo, K, Nowarski, R, Gross, J.D, Kotler, M, Harris, R.S, Matsuo, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model
J.Mol.Biol., 389, 2009
|
|
