1QM9
 
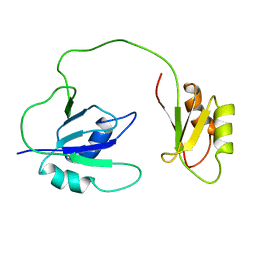 | | NMR, REPRESENTATIVE STRUCTURE | | Descriptor: | POLYPYRIMIDINE TRACT-BINDING PROTEIN | | Authors: | Conte, M.R, Grune, T, Curry, S, Matthews, S. | | Deposit date: | 1999-09-22 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of Tandem RNA Recognition Motifs from Polypyrimidine Tract Binding Protein Reveals Novel Features of the Rrm Fold
Embo J., 19, 2000
|
|
1QMC
 
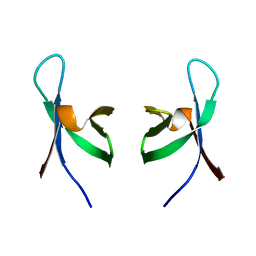 | | C-terminal DNA-binding domain of HIV-1 integrase, NMR, 42 structures | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Eijkelenboom, A.P.A.M, Sprangers, R, Hard, K, Puras Lutzke, R.A, Plasterk, R.H.A, Boelens, R, Kaptein, R. | | Deposit date: | 1999-09-27 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the C-Terminal DNA-Binding Domain of Human Immunovirus-1 Integrase.
Proteins: Struct.,Funct., Genet., 36, 1999
|
|
1QMS
 
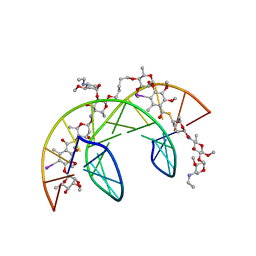 | | Head-to-Tail Dimer of Calicheamicin gamma-1-I Oligosaccharide Bound to DNA Duplex, NMR, 9 Structures | | Descriptor: | CALICHEAMICIN GAMMA-1-OLIGOSACCHARIDE, DNA (5'-D(*GP*CP*AP*CP*CP*TP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*AP*GP*GP*TP*GP*C)-3'), ... | | Authors: | Bifulco, G, Galeone, A, Nicolaou, K.C, Chazin, W.J, Gomez-Paloma, L. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Complex between the Head-to-Tail Dimer of Calicheamicin Gamma-1-I Oligosaccharide and a DNA Duplex Containing D(ACCT) and D(TCCT) High-Affinity Binding Sites
J.Am.Chem.Soc., 120, 1998
|
|
1QMW
 
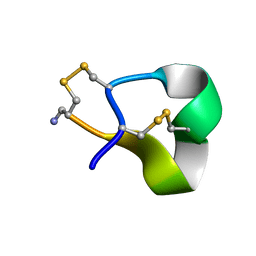 | | Solution structure of alpha-conotoxin SI | | Descriptor: | ALPHA-CONOTOXIN SI | | Authors: | Benie, A.J, Whitford, D, Hargittai, B, Barany, G, Janes, R.W. | | Deposit date: | 1999-10-08 | | Release date: | 2000-08-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha-Conotoxin Si
FEBS Lett., 476, 2000
|
|
1QN0
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QND
 
 | | STEROL CARRIER PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Lopez-Garcia, F, Szyperski, T, Dyer, J.H, Choinowski, T, Seedorf, U, Hauser, H, Wuthrich, K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Sterol Carrier Protein-2: Implications for the Biological Role
J.Mol.Biol., 295, 2000
|
|
1QNK
 
 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
1QNZ
 
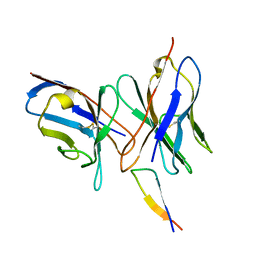 | | NMR structure of the 0.5b anti-HIV antibody complex with the gp120 V3 peptide | | Descriptor: | 0.5B ANTIBODY (HEAVY CHAIN), 0.5B ANTIBODY (LIGHT CHAIN), GP120 | | Authors: | Tugarinov, V, Zvi, A, Levy, R, Hayek, Y, Matsushita, S, Anglister, J. | | Deposit date: | 1999-10-26 | | Release date: | 2000-06-03 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Anti-Gp120 Antibody Complex with a V3 Peptide Reveals a Surface Important for Co-Receptor Binding
Structure, 8, 2000
|
|
1QO6
 
 | |
1QP6
 
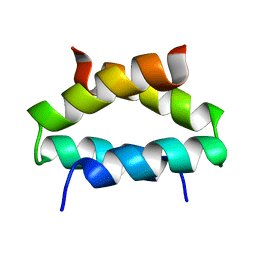 | | SOLUTION STRUCTURE OF ALPHA2D | | Descriptor: | PROTEIN (ALPHA2D) | | Authors: | Hill, R.B, DeGrado, W.F. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha2D, A Nativelike De Novo Designed Protein
J.Am.Chem.Soc., 120, 1998
|
|
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
1QQ3
 
 | | THE SOLUTION STRUCTURE OF THE HEME BINDING VARIANT ARG98CYS OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, HEME B/C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Ciofi-Baffoni, S, Barker, P.D, Woodyear, T. | | Deposit date: | 1999-06-10 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562.
Biochemistry, 39, 2000
|
|
1QQI
 
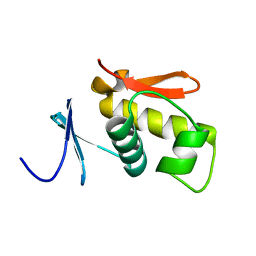 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
1QQV
 
 | |
1QRJ
 
 | | Solution structure of htlv-i capsid protein | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Khorasanizadeh, S, Campos-Olivas, R, Clark, C.A, Summers, M.F. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein.
J.Biomol.NMR, 14, 1999
|
|
1QSK
 
 | | NMR-DERIVED SOLUTION STRUCTURE OF A FIVE-ADENINE BULGE LOOP WITHIN A DNA DUPLEX | | Descriptor: | 5'-D(CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*CP)-3', 5'-D(GP*CP*AP*TP*CP*GP*AP*AP*AP*AP*AP*GP*CP*TP*AP*CP*GP)-3' | | Authors: | Dornberger, U, Hillisch, A, Gollmick, F, Fritzsche, H, Diekmann, S. | | Deposit date: | 1999-06-22 | | Release date: | 1999-11-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a five-adenine bulge loop within a DNA duplex.
Biochemistry, 38, 1999
|
|
1QTG
 
 | | AVERAGED NMR MODEL OF SWITCH ARC, A DOUBLE MUTANT OF ARC REPRESSOR | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H.J, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 1999-06-27 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolution of a protein fold in vitro.
Science, 284, 1999
|
|
1QU5
 
 | |
1QUZ
 
 | | Solution structure of the potassium channel scorpion toxin HSTX1 | | Descriptor: | HSTX1 TOXIN | | Authors: | Savarin, P, Romi-Lebrun, R, Zinn-Justin, S, Lebrun, B, Nakajima, T, Gilquin, B, Menez, A. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional consequences of the presence of a fourth disulfide bridge in the scorpion short toxins: solution structure of the potassium channel inhibitor HsTX1.
Protein Sci., 8, 1999
|
|
1QVK
 
 | | Structure of the antimicrobial hexapeptide cyc-(RRWWRF) bound to DPC micelles | | Descriptor: | c-RW | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2003-08-28 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antimicrobial, Cationic Hexapeptide Cyclo(RRWWRF) and Its Analogues in Solution and Bound to Detergent Micelles
Chembiochem, 6, 2005
|
|
1QVL
 
 | | Structure of the antimicrobial hexapeptide cyc-(RRWWRF) bound to SDS micelles | | Descriptor: | c-RW | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2003-08-28 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antimicrobial, Cationic Hexapeptide Cyclo(RRWWRF) and Its Analogues in Solution and Bound to Detergent Micelles
Chembiochem, 6, 2005
|
|
1QVP
 
 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
1QWA
 
 | | NMR structure of 5'-r(GGAUGCCUCCCGAGUGCAUCC): an RNA hairpin derived from the mouse 5'ETS that binds nucleolin RBD12. | | Descriptor: | 18S ribosomal RNA, 5'ETS | | Authors: | Finger, L.D, Trantirek, L, Johansson, C, Feigon, J. | | Deposit date: | 2003-09-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of Stem-loop RNAs that Bind to the Two N-terminal RNA Binding Domains of Nucleolin
Nucleic Acids Res., 31, 2003
|
|
