8QKX
 
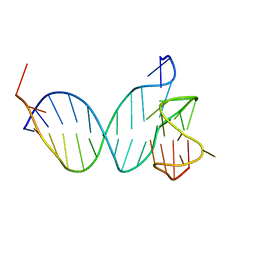 | |
8QNJ
 
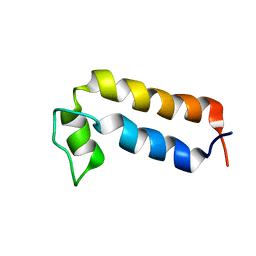 | |
8QNT
 
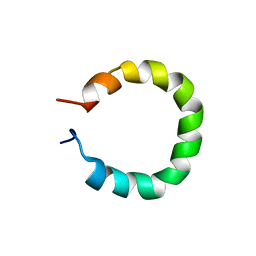 | |
8QNV
 
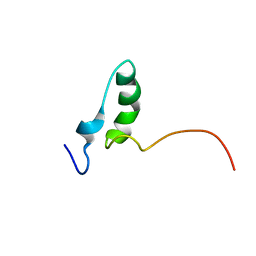 | |
8QOR
 
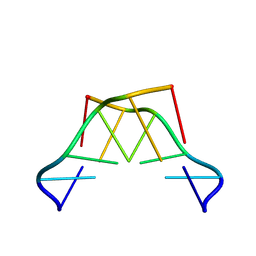 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QPD
 
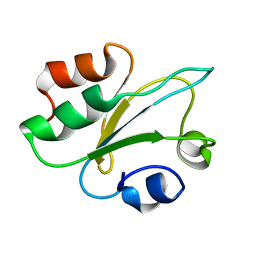 | | Structure of thioredoxin m from pea | | Descriptor: | Thioredoxin M-type, chloroplastic | | Authors: | Neira, J.L, Camara Artigas, A. | | Deposit date: | 2023-10-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure, dynamics and binding of thioredoxin m from Pisum sativum.
Int.J.Biol.Macromol., 262, 2024
|
|
8QPY
 
 | |
8QRX
 
 | |
8QSX
 
 | |
8QTQ
 
 | | Thermostable WW domain | | Descriptor: | WW domain | | Authors: | Kovermann, M, Thomas, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Thermostable WW-Domain Scaffold to Design Functional beta-Sheet Miniproteins.
J.Am.Chem.Soc., 2024
|
|
8QWQ
 
 | |
8QXI
 
 | | SipA solution structure | | Descriptor: | Uncharacterized protein SEF0032 | | Authors: | Neira, J.L. | | Deposit date: | 2023-10-24 | | Release date: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the cyanobacterial regulator SipA.
Arch.Biochem.Biophys., 754, 2024
|
|
8R1X
 
 | |
8R4E
 
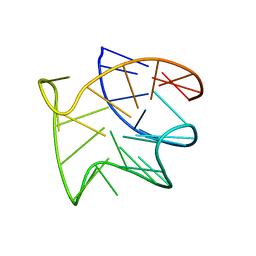 | |
8R4W
 
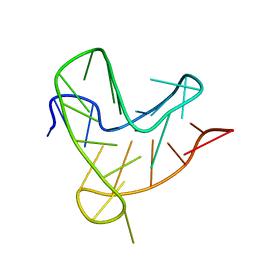 | |
8R62
 
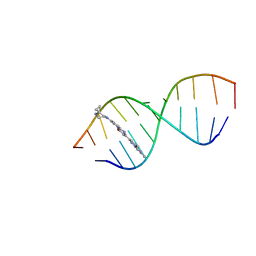 | | Solution structure of Risdiplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 7-(4,7-diazaspiro[2.5]octan-7-yl)-2-(2,8-dimethylimidazo[1,2-b]pyridazin-6-yl)-1~{H}-pyrido[1,2-a]pyrimidin-4-one, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8R63
 
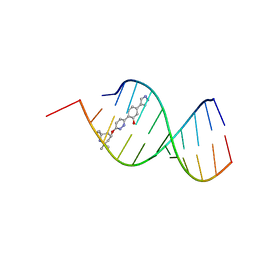 | | Solution structure of branaplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 5-(1~{H}-pyrazol-4-yl)-2-[6-(2,2,6,6-tetramethylpiperidin-4-yl)oxypyridazin-3-yl]phenol, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8R6D
 
 | |
8R6G
 
 | |
8R6H
 
 | |
8R6T
 
 | | NMR solution structure of thyropin IrThy-Cd from the hard tick Ixodes ricinus | | Descriptor: | Putative two thyropin protein (Fragment) | | Authors: | Srb, P, Veverka, V, Matouskova, Z, Orsaghova, K, Mares, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | An Unusual Two-Domain Thyropin from Tick Saliva: NMR Solution Structure and Highly Selective Inhibition of Cysteine Cathepsins Modulated by Glycosaminoglycans.
Int J Mol Sci, 25, 2024
|
|
8R8P
 
 | |
8RAJ
 
 | | NMR structure of PKS docking domains | | Descriptor: | Beta-ketoacyl synthase, Trimethylamine monooxygenase | | Authors: | Scat, S, Weissman, K.J, Chagot, B. | | Deposit date: | 2023-12-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | SOLUTION NMR | | Cite: | Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Rsc Chem Biol, 5, 2024
|
|
8RD6
 
 | |
8RHS
 
 | |
