6ES6
 
 | |
6ES7
 
 | |
6ESP
 
 | |
6EVI
 
 | | solution NMR structure of EB1 C terminus (191-260) | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Barsukov, I.L, Almeida, T.B. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting SxIP-EB1 interaction: An integrated approach to the discovery of small molecule modulators of dynamic binding sites.
Sci Rep, 7, 2017
|
|
6EVQ
 
 | |
6EWS
 
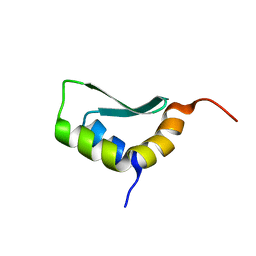 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWT
 
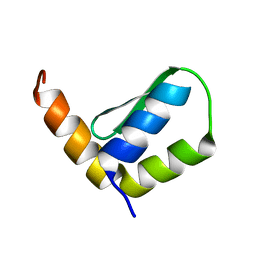 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWU
 
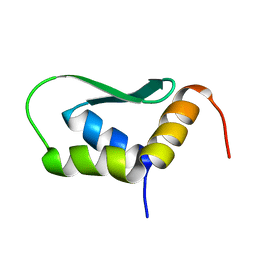 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
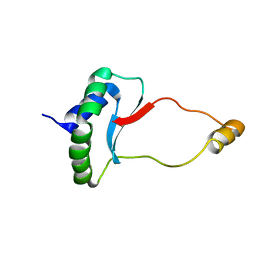 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EY3
 
 | |
6EZ0
 
 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
6EZ4
 
 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
6F0Y
 
 | | Rtt109 peptide bound to Asf1 | | Descriptor: | Histone chaperone ASF1, histone acetyltransferase Rtt109 C-terminus | | Authors: | Lercher, L, Kirkpatrick, J.P, Carlomagno, T. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the Asf1-Rtt109 interaction and its role in histone acetylation.
Nucleic Acids Res., 46, 2018
|
|
6F24
 
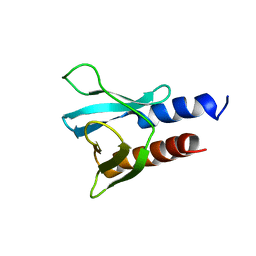 | | PH domain from PfAPH | | Descriptor: | C-terminal PH domain from PfAPH | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | C-terminal PH domain from P. falciparum acylated plekstrin homology domain containing protein (APH)
To Be Published
|
|
6F27
 
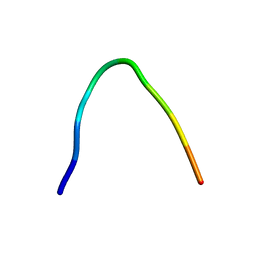 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F2X
 
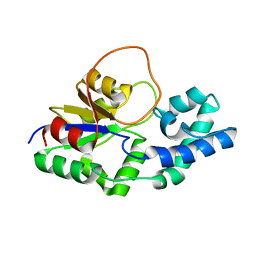 | | Structural characterization of the Mycobacterium tuberculosis Protein Tyrosine Kinase A (PtkA) | | Descriptor: | Protein Tyrosine Kinase A | | Authors: | Niesteruk, A, Jonker, H.R.A, Sreeramulu, S, Richter, C, Hutchison, M, Linhard, V, Schwalbe, H. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The domain architecture of PtkA, the first tyrosine kinase fromMycobacterium tuberculosis, differs from the conventional kinase architecture.
J. Biol. Chem., 293, 2018
|
|
6F3K
 
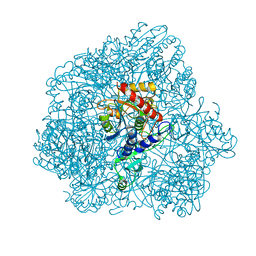 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F46
 
 | |
6F4Z
 
 | |
6F55
 
 | | Complex structure of PACSIN SH3 domain and TRPV4 proline rich region | | Descriptor: | PACSIN 3, PRR | | Authors: | Glogowski, N.A, Goretzki, B, Diehl, E, Duchardt-Ferner, E, Hacker, C, Hellmich, U.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Structure, 26, 2018
|
|
6F61
 
 | |
