2LON
 
 | |
2LOO
 
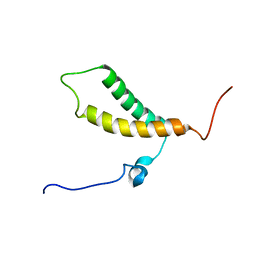
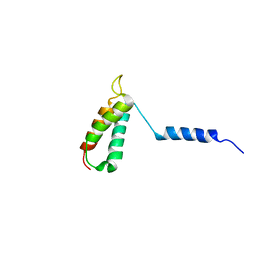 | | Backbone structure of human membrane protein TMEM14A from NOE data | | Descriptor: | Transmembrane protein 14A | | Authors: | Eichmann, C, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOP
 
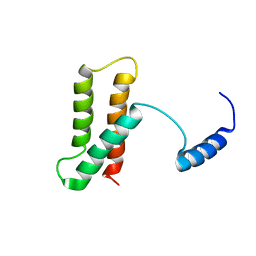 | | Backbone structure of human membrane protein TMEM14A | | Descriptor: | Transmembrane protein 14A | | Authors: | Eichmann, C, Klammt, C, Maslennikov, I, Kwiatkowski, W, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOQ
 
 | | Backbone structure of human membrane protein FAM14B (Interferon alpha-inducible protein 27-like protein 1) | | Descriptor: | Interferon alpha-inducible protein 27-like protein 1 | | Authors: | Klammt, C, Chui, E.J, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOR
 
 | | Backbone structure of human membrane protein TMEM141 | | Descriptor: | Transmembrane protein 141 | | Authors: | Bayrhuber, M, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOS
 
 | | Backbone structure of human membrane protein TMEM14C | | Descriptor: | Transmembrane protein 14C | | Authors: | Klammt, C, Vajpai, N, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOT
 
 | | AR55 solubilised in SDS micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
2LOU
 
 | | AR55 solubilised in DPC micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural features of the apelin receptor N-terminal tail and first transmembrane segment implicated in ligand binding and receptor trafficking.
Biochim.Biophys.Acta, 1828, 2013
|
|
2LOV
 
 | | AR55 solubilised in LPPG micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
2LOW
 
 | | Solution structure of AR55 in 50% HFIP | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
2LOX
 
 | |
2LOY
 
 | | Refined Miminal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from Caenorhabditis elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Lee, H, Janjua, H, Maglaqui, M, Cooper, B, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOZ
 
 | | The novel binding mode of DLC1 and Tensin2 PTB domain | | Descriptor: | Rho GTPase-activating protein 7, Tensin-like C1 domain-containing phosphatase | | Authors: | Liu, C, Chen, L, Zhu, G. | | Deposit date: | 2012-01-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphotyrosine binding (PTB) domain of human tensin2 protein in complex with deleted in liver cancer 1 (DLC1) peptide reveals a novel peptide binding mode.
J.Biol.Chem., 287, 2012
|
|
2LP0
 
 | |
2LP1
 
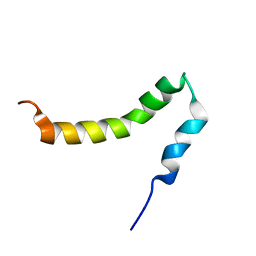 | | The solution NMR structure of the transmembrane C-terminal domain of the amyloid precursor protein (C99) | | Descriptor: | C99 | | Authors: | Barrett, P.J, Song, Y, Van Horn, W.D, Hustedt, E.J, Schafer, J.M, Hadziselimovic, A, Beel, A.J, Sanders, C.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol.
Science, 336, 2012
|
|
2LP2
 
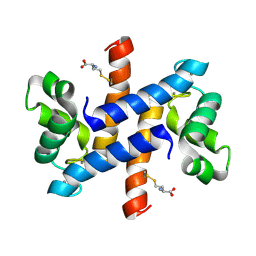 | | Solution structure and dynamics of human S100A1 protein modified at cysteine 85 with homocysteine disulfide bond formation in calcium saturated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, CALCIUM ION, Protein S100-A1 | | Authors: | Nowakowski, M.E, Jaremko, L, Jaremko, M, Zdanowski, K, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2LP3
 
 | | Solution structure of S100A1 Ca2+ | | Descriptor: | CALCIUM ION, Protein S100-A1 | | Authors: | Budzinska, M, Ruszczynska-Bartnik, K, Belczyk-Ciesielska, A, Bierzynski, A, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2LP4
 
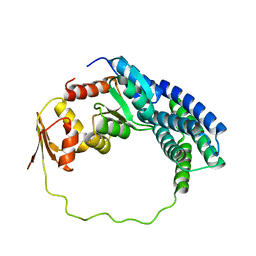 | | Solution structure of P1-CheY/P2 complex in bacterial chemotaxis | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheY | | Authors: | Dahlquist, F, Mo, G, Zhou, H, Kamamura, T. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a complex of the histidine autokinase CheA with its substrate CheY.
Biochemistry, 51, 2012
|
|
2LP5
 
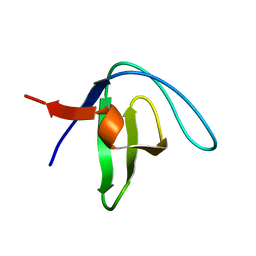 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2LP6
 
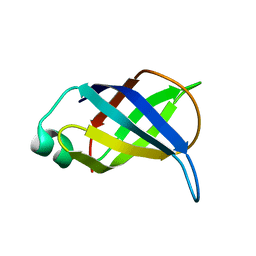 | | Refined Solution NMR Structure of the 50S ribosomal protein L35Ae from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target (NESG) PfR48 | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Yu, B, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-02 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus.
Proteins, 80, 2012
|
|
2LP7
 
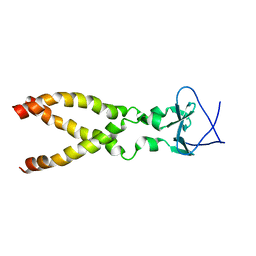 | |
2LP8
 
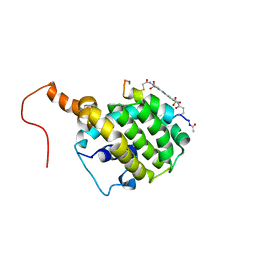 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
2LP9
 
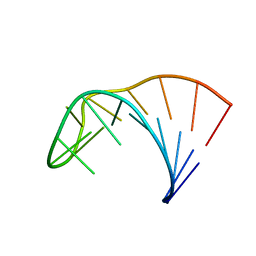 | | Pseudo-triloop from the sub-genomic promoter of Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*AP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
2LPA
 
 | | Mutant of the sub-genomic promoter from Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
2LPB
 
 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
