2K7G
 
 | | Solution Structure of varv F | | Descriptor: | Varv peptide F | | Authors: | Wang, C.K. | | Deposit date: | 2008-08-10 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold
J.Biol.Chem., 284, 2009
|
|
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | Descriptor: | Stress-induced protein SAM22 | | Authors: | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
2K7I
 
 | | Solution NMR structure of protein ATU0232 from AGROBACTERIUM TUMEFACIENS. Northeast Structural Genomics Consortium (NESG) target AtT3. Ontario Center for Structural Proteomics target ATC0223. | | Descriptor: | UPF0339 protein Atu0232 | | Authors: | Lemak, A, Srisailam, S, Yee, A, Bansal, S, Semesi, A, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATU0232 from Agrobacterium Tumefaciens.
To be Published
|
|
2K7J
 
 | | Human Acylphosphatase(AcPh) surface charge-optimized | | Descriptor: | Acylphosphatase-1 | | Authors: | Gribenko, A.V, Patel, M.M, Liu, J, McCallum, S.A, Wang, C, Makhatadze, G.I. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rational stabilization of enzymes by computational redesign of surface charge-charge interactions
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K7K
 
 | | Human Acylphosphatase (AcPh) common type | | Descriptor: | Acylphosphatase-1 | | Authors: | Gribenko, A.V, Patel, M.M, Liu, J, McCallum, S.A, Wang, C, Makhatadze, G.I. | | Deposit date: | 2008-08-13 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rational stabilization of enzymes by computational redesign of surface charge-charge interactions
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K7L
 
 | | NMR structure of a complex formed by the C-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 | | Descriptor: | General transcription factor IIF subunit 1, centFCP1-T584PO4 peptide | | Authors: | Yang, A, Abbott, K.L, Desjardins, A, Di Lello, P, Omichinski, J.G, Legault, P. | | Deposit date: | 2008-08-13 | | Release date: | 2009-06-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex formed by the carboxyl-terminal domain of human RAP74 and a phosphorylated peptide from the central domain of the FCP1 phosphatase
Biochemistry, 48, 2009
|
|
2K7M
 
 | | Structure of the Connexin40 Carboxyl terminal Domain | | Descriptor: | Gap junction alpha-5 protein | | Authors: | Bouvier, D, Spagnol, G, Kieken, F, Vitrac, H, Kellezi, A, Forge, V. | | Deposit date: | 2008-08-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Characterization of the structure and intermolecular interactions between the connexin40 and connexin43 carboxyl-terminal and cytoplasmic loop domains.
J.Biol.Chem., 284, 2009
|
|
2K7N
 
 | |
2K7O
 
 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
2K7P
 
 | | Filamin A Ig-like domains 16-17 | | Descriptor: | Filamin-A | | Authors: | Heikkinen, O.K, Kilpelainen, I, Koskela, H, Permi, P, Heikkinen, S, Ylanne, J. | | Deposit date: | 2008-08-19 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin
J.Biol.Chem., 284, 2009
|
|
2K7Q
 
 | | Filamin A Ig-like domains 18-19 | | Descriptor: | Filamin-A | | Authors: | Heikkinen, O.K, Kilpelainen, I, Koskela, H, Permi, P, Heikkinen, S, Ylanne, J. | | Deposit date: | 2008-08-19 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin
J.Biol.Chem., 284, 2009
|
|
2K7R
 
 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
2K7S
 
 | |
2K7V
 
 | |
2K7W
 
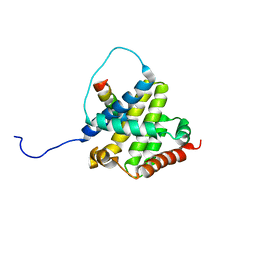 | | BAX Activation is Initiated at a Novel Interaction Site | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Gavathiotis, E, Suzuki, M, Davis, M.L, Pitter, K, Bird, G.H, Katz, S.G, Tu, H.C, Kim, H, Cheng, E.H, Tjandra, N, Walensky, L.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAX activation is initiated at a novel interaction site.
Nature, 455, 2008
|
|
2K7X
 
 | |
2K7Y
 
 | |
2K7Z
 
 | |
2K84
 
 | | Solution Structure of the equine infectious anemia virus p9 GAG protein | | Descriptor: | P9 | | Authors: | Sharma, A, Bruns, K, Roeder, R, Henklein, P, Votteler, J, Wray, V, Sharma, U. | | Deposit date: | 2008-09-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Equine Infectious Anemia Virus p9 protein: a rationalization of its different ALIX binding requirements compared to the analogous HIV-p6 protein
Bmc Struct.Biol., 9, 2009
|
|
2K85
 
 | | p190-A RhoGAP FF1 domain | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1 | | Authors: | Bonet, R, Ruiz, L, Martin-Malpartida, P, Macias, M. | | Deposit date: | 2008-09-02 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on human p190-A RhoGAPFF1 revealed that domain phosphorylation by the PDGF-receptor alpha requires its previous unfolding.
J.Mol.Biol., 389, 2009
|
|
2K86
 
 | | Solution Structure of FOXO3a Forkhead domain | | Descriptor: | Forkhead box protein O3 | | Authors: | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | Deposit date: | 2008-09-02 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
2K87
 
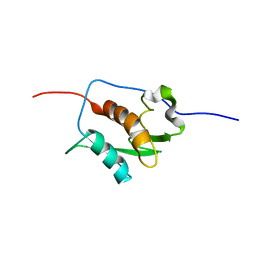 | | NMR STRUCTURE OF A PUTATIVE RNA BINDING PROTEIN (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | Non-structural protein 3 of Replicase polyprotein 1a | | Authors: | Serrano, P, Wuthrich, K, Johnson, M.A, Chatterjee, A, Wilson, I, Pedrini, B.F, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3.
J.Virol., 83, 2009
|
|
2K88
 
 | | Association of subunit d (Vma6p) and E (Vma4p) with G (Vma10p) and the NMR solution structure of subunit G (G1-59) of the Saccharomyces cerevisiae V1VO ATPase | | Descriptor: | Vacuolar proton pump subunit G | | Authors: | Sankaranarayanan, N, Gayen, S, Thaker, Y, Subramanian, V, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Assembly of subunit d (Vma6p) and G (Vma10p) and the NMR solution structure of subunit G (G(1-59)) of the Saccharomyces cerevisiae V(1)V(O) ATPase.
Biochim.Biophys.Acta, 1787, 2009
|
|
2K89
 
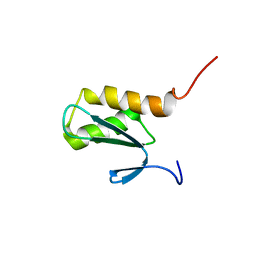 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 cis isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2K8A
 
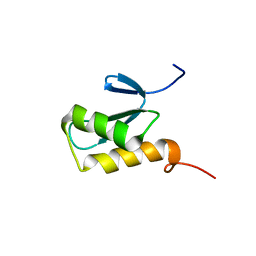 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 trans isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
