1EYO
 
 | |
1EZA
 
 | |
1EZO
 
 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | 分子名称: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | 著者 | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | 登録日 | 2000-05-11 | | 公開日 | 2001-05-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1EZP
 
 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN USING PEPTIDE ORIENTATIONS FROM DIPOLAR COUPLINGS | | 分子名称: | MALTODEXTRIN BINDING PERIPLASMIC PROTEIN | | 著者 | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | 登録日 | 2000-05-11 | | 公開日 | 2001-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-11 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-12 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1F03
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | 分子名称: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | 登録日 | 2000-05-14 | | 公開日 | 2000-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | 分子名称: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | 登録日 | 2000-05-14 | | 公開日 | 2000-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
1F0Z
 
 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | 分子名称: | THIS PROTEIN | | 著者 | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | 登録日 | 2000-05-17 | | 公開日 | 2001-01-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
1F16
 
 | |
1F22
 
 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS. COMPARISON BETWEEN THE REDUCED AND THE OXIDIZED FORMS. | | 分子名称: | CYTOCHROME C7, HEME C | | 著者 | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | 登録日 | 2000-05-23 | | 公開日 | 2000-06-21 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1F2G
 
 | | THE NMR SOLUTION STRUCTURE OF THE 3FE FERREDOXIN II FROM DESULFOVIBRIO GIGAS, 15 STRUCTURES | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN II | | 著者 | Goodfellow, B.J, Macedo, A.L, Rodrigues, P, Wray, V, Moura, I, Moura, J.J.G. | | 登録日 | 1998-10-08 | | 公開日 | 1999-09-02 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a [3Fe-4S] ferredoxin: oxidised ferredoxin II from Desulfovibrio gigas.
J.Biol.Inorg.Chem., 4, 1999
|
|
1F2H
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE TNFR1 ASSOCIATED PROTEIN, TRADD. | | 分子名称: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN | | 著者 | Tsao, D, McDonaugh, T, Malakian, K, Xu, G.-Y, Telliez, J.-B, Hsu, H, Lin, L.-L. | | 登録日 | 2000-05-24 | | 公開日 | 2001-05-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway.
Mol.Cell, 5, 2000
|
|
1F2R
 
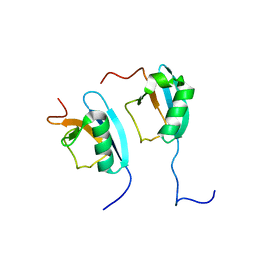 | | NMR STRUCTURE OF THE HETERODIMERIC COMPLEX BETWEEN CAD DOMAINS OF CAD AND ICAD | | 分子名称: | CASPASE-ACTIVATED DNASE, INHIBITOR OF CASPASE-ACTIVATED DNASE | | 著者 | Otomo, T, Sakahira, H, Uegaki, K, Nagata, S, Yamazaki, T. | | 登録日 | 2000-05-29 | | 公開日 | 2000-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the heterodimeric complex between CAD domains of CAD and ICAD.
Nat.Struct.Biol., 7, 2000
|
|
1F3C
 
 | |
1F3R
 
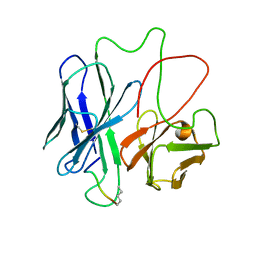 | | COMPLEX BETWEEN FV ANTIBODY FRAGMENT AND AN ANALOGUE OF THE MAIN IMMUNOGENIC REGION OF THE ACETYLCHOLINE RECEPTOR | | 分子名称: | ACETYLCHOLINE RECEPTOR ALPHA, FV ANTIBODY FRAGMENT | | 著者 | Kleinjung, J, Petit, M.-C, Orlewski, P, Mamalaki, A, Tzartos, S.-J, Tsikaris, V, Sakarellos-Daitsiotis, M, Sakarellos, C, Marraud, M, Cung, M.-T. | | 登録日 | 2000-06-06 | | 公開日 | 2000-06-15 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The third-dimensional structure of the complex between an Fv antibody fragment and an analogue of the main immunogenic region of the acetylcholine receptor: a combined two-dimensional NMR, homology, and molecular modeling approach.
Biopolymers, 53, 2000
|
|
1F3Y
 
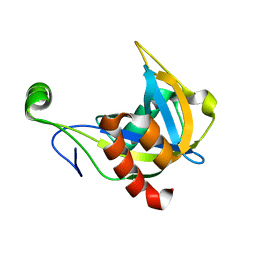 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | 分子名称: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | 著者 | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | 登録日 | 2000-06-06 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
1F40
 
 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | 分子名称: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | 著者 | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | 登録日 | 2000-06-07 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1F43
 
 | | SOLUTION STRUCTURE OF THE MATA1 HOMEODOMAIN | | 分子名称: | MATING-TYPE PROTEIN A-1 | | 著者 | Anderson, J.S, Forman, M, Modleski, S, Dahlquist, F.W, Baxter, S.M. | | 登録日 | 2000-06-07 | | 公開日 | 2000-07-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cooperative ordering in homeodomain-DNA recognition: solution structure and dynamics of the MATa1 homeodomain.
Biochemistry, 39, 2000
|
|
1F53
 
 | | NMR STRUCTURE OF KILLER TOXIN-LIKE PROTEIN SKLP | | 分子名称: | YEAST KILLER TOXIN-LIKE PROTEIN | | 著者 | Ohki, S, Kariya, E, Hiraga, K, Wakamiya, A, Isobe, T, Oda, K, Kainosho, M. | | 登録日 | 2000-06-12 | | 公開日 | 2000-12-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of Streptomyces killer toxin-like protein, SKLP: further evidence for the wide distribution of single-domain betagamma-crystallin superfamily proteins.
J.Mol.Biol., 305, 2001
|
|
1F5G
 
 | |
1F5H
 
 | |
1F5U
 
 | |
1F5Y
 
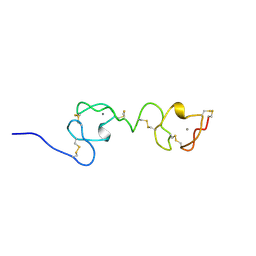 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | 分子名称: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | 著者 | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | 登録日 | 2000-06-18 | | 公開日 | 2000-08-30 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
1F6V
 
 | |
