2MES
 
 | |
2MET
 
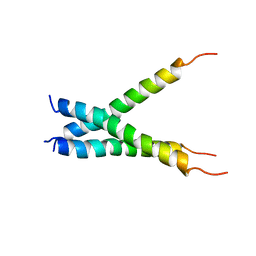 | | NMR spatial structure of the trimeric mutant TM domain of VEGFR2 receptor. | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.A, Shulepko, M, Lyukmanova, E.N, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
2MEU
 
 | | NMR spatial structure of mutant dimeric TM domain of VEGFR2 receptor | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.S, Lyukmanova, E.N, Shulepko, M.A, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
2MEW
 
 | |
2MEX
 
 | | Structure of the tetrameric building block of the Salmonella Typhimurium PrgI Type three secretion system needle | | Descriptor: | Protein PrgI | | Authors: | Loquet, A, Habenstein, B, Chevelkov, V, Giller, K, Becker, S, Lange, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and handedness of the building block of a biological assembly.
J.Am.Chem.Soc., 135, 2013
|
|
2MEY
 
 | |
2MEZ
 
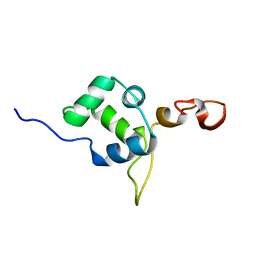 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
2MF0
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF2
 
 | |
2MF3
 
 | | SGTX-Sf1a | | Descriptor: | U2-segestritoxin-Sf1a | | Authors: | Mobli, M, Bende, N.S, King, G.F. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The insecticidal spider toxin SFI1 is a knottin peptide that blocks the pore of insect voltage-gated sodium channels via a large beta-hairpin loop.
Febs J., 282, 2015
|
|
2MF4
 
 | | 1H, 13C, 15N chemical shift assignments of Streptomyces virginiae VirA acp5a | | Descriptor: | Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Mazon, H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR assignements of ACP5a
To be Published
|
|
2MF6
 
 | | Solution NMR structure of Chimeric Avidin, ChiAVD(I117Y), in the biotin bound form | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Tossavainen, H, Kukkurainen, S, Maatta, J.A.E, Pihlajamaa, T, Hytonen, V.P, Kulomaa, M.S, Permi, P. | | Deposit date: | 2013-10-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chimeric Avidin - NMR Structure and Dynamics of a 56 kDa Homotetrameric Thermostable Protein
Plos One, 9, 2014
|
|
2MF7
 
 | | Solution structure of the ims domain of the mitochondrial import protein TIM21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bajaj, R, Jaremko, L, Jaremko, M, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space.
Structure, 22, 2014
|
|
2MF8
 
 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
2MF9
 
 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
2MFA
 
 | | Mambalgin-2 | | Descriptor: | Mambalgin-2 | | Authors: | Schroeder, C.I, Rash, L.D, Vila-Farres, X, Rosengren, K.J, Mobli, M, King, G.F, Alewood, P.F, Craik, D.J, Durek, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2MFC
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFD
 
 | |
2MFE
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFF
 
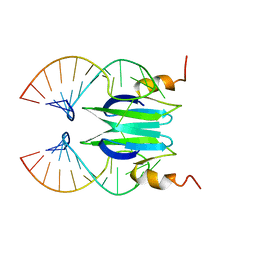 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFG
 
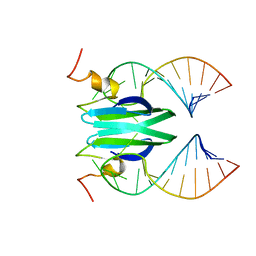 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
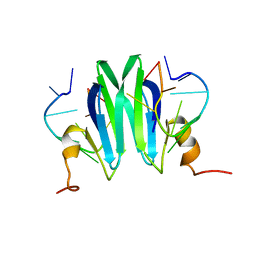 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFI
 
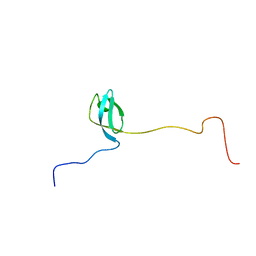 | | Domain 1 of E. coli ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Giraud, P, Crechet, J, Bontems, F, Uzan, M, Sizun, C. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Resonance assignment of the ribosome binding domain of E. coli ribosomal protein S1.
Biomol.Nmr Assign., 9, 2015
|
|
2MFJ
 
 | |
