9C61
 
 | | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26 | | Descriptor: | (4R)-4-[4-(5-fluoro-1H-indol-3-yl)piperidine-1-carbonyl]piperidin-2-one, Non-specific serine/threonine protein kinase, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Kutera, M, Ilyassov, O, Seitova, A, Loppnau, P, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-06-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26
To be published
|
|
9C6P
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | 3-chloro-N-(5-chloro-2-methyl-1,3-benzothiazol-6-yl)-2-hydroxybenzene-1-sulfonamide, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C81
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (2R)-2-phenoxy-3-{[(1S,2S,4S)-spiro[bicyclo[2.2.1]heptane-7,1'-cyclopropane]-2-carbonyl]amino}propanoic acid, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K, Bassim, V. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C82
 
 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
9C83
 
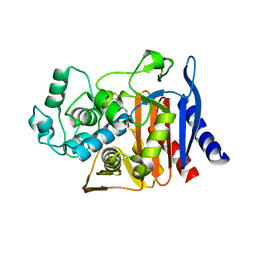 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | AmpC Beta-lactamase, N-[(3M)-3-(5-chloro-1,2,3-thiadiazol-4-yl)phenyl]-5-methyl-3-oxo-2,3-dihydro-1,2-oxazole-4-sulfonamide | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C84
 
 | |
9C8J
 
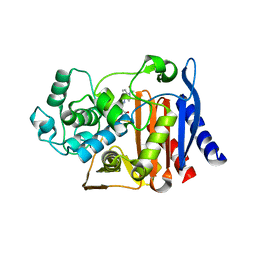 | |
9EMA
 
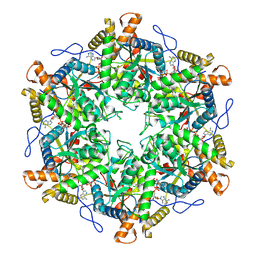 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EN6
 
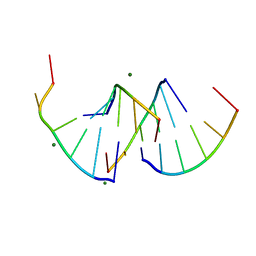 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
9ENZ
 
 | |
9EO0
 
 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
9EO2
 
 | |
9EO5
 
 | |
9EO8
 
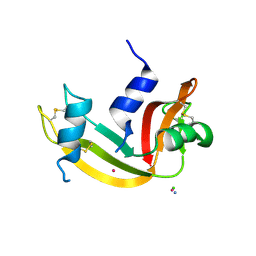 | |
9EOJ
 
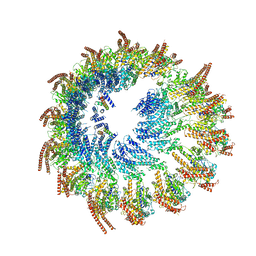 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
9EOU
 
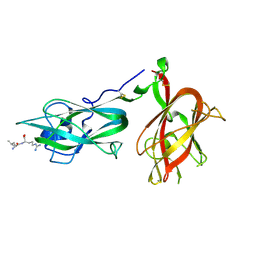 | |
9EPV
 
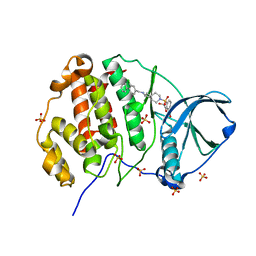 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3-chloranyl-4-phenyl-phenyl)methylamino]-7-azaspiro[3.5]nonan-7-yl]sulfonyl]-1,3-dimethyl-benzimidazol-2-one, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333
To Be Published
|
|
9EPW
 
 | |
9EPX
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331 | | Descriptor: | 1,2-ETHANEDIOL, 7-[(2-chloranyl-1,3-benzothiazol-6-yl)sulfonyl]-~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-7-azaspiro[3.5]nonan-2-amine, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3331
To Be Published
|
|
9EPY
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330
To Be Published
|
|
9EPZ
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3337 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3337
To Be Published
|
|
9EQ0
 
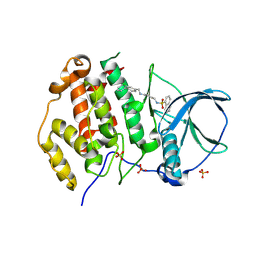 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJG12 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[4-[(3-chloranyl-4-phenyl-phenyl)methylamino]butyl]isoquinoline-5-sulfonamide | | Authors: | Kraemer, A, Greco, F, Gerninghaus, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJG12
To Be Published
|
|
9EQ1
 
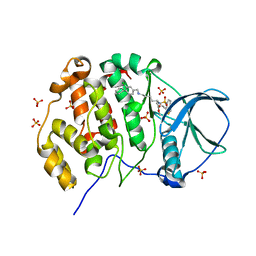 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJM24 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Moeckel, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJM24
To Be Published
|
|
9EQ3
 
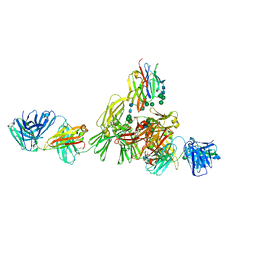 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 8
To be published
|
|
9EQ4
 
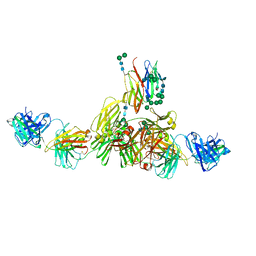 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5
To be published
|
|






