7A5H
 
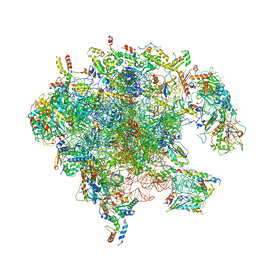 | | Structure of the split human mitoribosomal large subunit with rescue factors mtRF-R and MTRES1 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5I
 
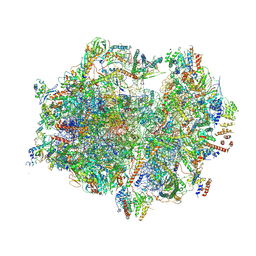 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5J
 
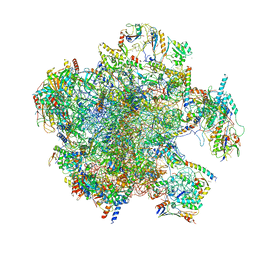 | | Structure of the split human mitoribosomal large subunit with P-and E-site mt-tRNAs | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5K
 
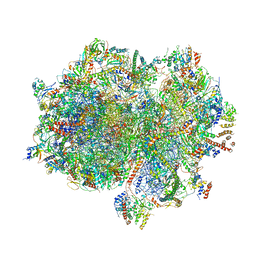 | | Structure of the human mitoribosome in the post translocation state bound to mtEF-G1 | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7A6X
 
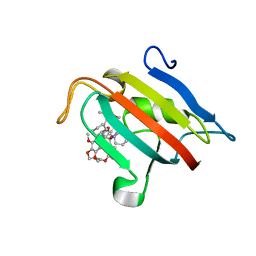 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | Descriptor: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7A7D
 
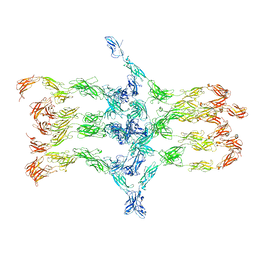 | | Cadherin fit into cryo-ET map | | Descriptor: | Desmocollin-2, Desmoglein-2 | | Authors: | Sikora, M, Ermel, U.H, Seybold, A, Kunz, M, Calloni, G, Reitz, J, Vabulas, R.M, Hummer, G, Frangakis, A.S. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A8W
 
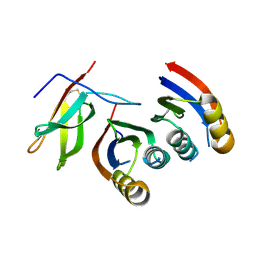 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with an engineered HMA domain of Pikp-1 from rice (Oryza sativa) | | Descriptor: | NBS-LRR class disease resistance protein, Uncharacterized protein | | Authors: | Maidment, J.H.R, De la Concepcion, J.C, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
7A8Y
 
 | |
7A9C
 
 | |
7A9G
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis with intermediate 2-acetyl-thiamine diphosphate | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, 2-ACETYL-THIAMINE DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
7A9H
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
7A9U
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 3-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)-N-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-8-yl)propanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-(but-3-yn-1-yl)-3H-diazirin-3-yl)-N-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-8-yl)propanamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | A Photoaffinity-Based Fragment-Screening Platform for Efficient Identification of Protein Ligands.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7A9Y
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | Cellular retinoic acid-binding protein 1, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7A9Z
 
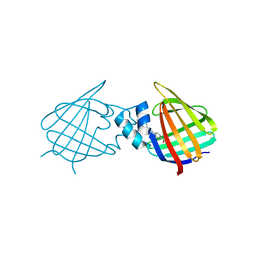 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 1 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AA0
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | (~{E})-3-[4-(4,4-dimethyl-1-propan-2-yl-2,3-dihydroquinolin-6-yl)phenyl]prop-2-enoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AA1
 
 | | Structural comparison of cellular retinoic acid binding proteins I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AAE
 
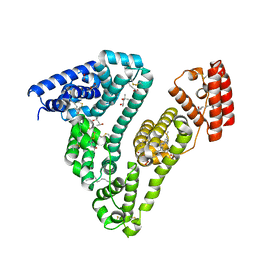 | | Crystal structure of Human serum albumin in complex with myristic acid at 2.27 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, FORMIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7AAI
 
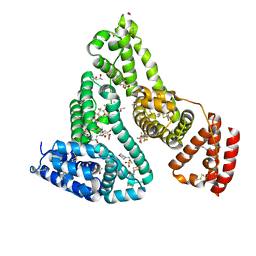 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
7ABF
 
 | | Human pre-Bact-1 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABG
 
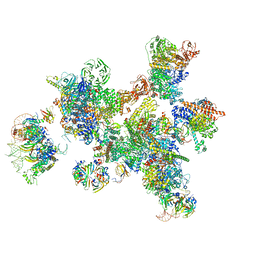 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7ABZ
 
 | | Structure of pre-accomodated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-08-18 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7AC0
 
 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
7AC7
 
 | | Structure of accomodated trans-translation complex on E. Coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-08-18 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7ACJ
 
 | | Structure of translocated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-11 | | Release date: | 2021-08-18 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|







