6Y01
 
 | |
6Y0C
 
 | | Influenza C virus polymerase in complex with human ANP32A - Subclass 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*GP*CP*CP*UP*UP*UP*U)-3'), ... | | Authors: | Fan, H, Carrique, L, Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6Y0G
 
 | | Structure of human ribosome in classical-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y0K
 
 | |
6Y0R
 
 | | Chitooligosaccharide oxidase | | Descriptor: | Chitooligosaccharide oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Savino, S, Fraaije, M.W. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Analysis of the structure and substrate scope of chitooligosaccharide oxidase reveals high affinity for C2-modified glucosamines.
Febs Lett., 594, 2020
|
|
6Y0S
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6Y25
 
 | | Streptavidin mutant S112R,K121E with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2I
 
 | |
6Y2J
 
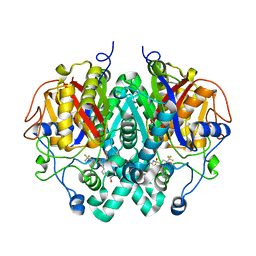 | | Crystal structure of M. tuberculosis KasA in complex with 4,4,4-trifluoro-N-(isoquinolin-6-yl)butane-1-sulfonamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, 4,4,4-tris(fluoranyl)-~{N}-isoquinolin-6-yl-butane-1-sulfonamide, SODIUM ION, ... | | Authors: | Chung, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Exploring the SAR of the beta-Ketoacyl-ACP Synthase Inhibitor GSK3011724A and Optimization around a Genotoxic Metabolite.
Acs Infect Dis., 6, 2020
|
|
6Y2L
 
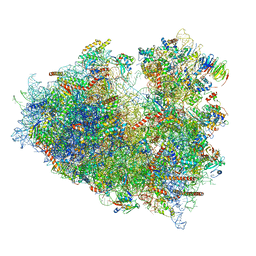 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y2M
 
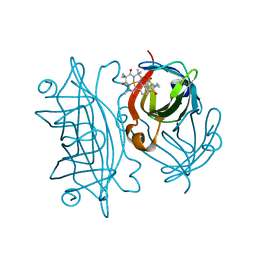 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2T
 
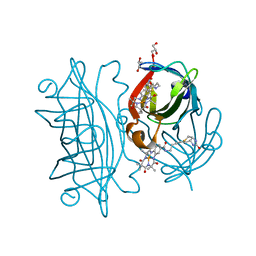 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y33
 
 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y34
 
 | | Streptavidin wildtype with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y3D
 
 | | X-ray structure of thermophilic C-phycocyanin from Galdiera phlegrea | | Descriptor: | ACETATE ION, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Ferraro, G, Lucignano, R, Marseglia, A, Merlino, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of C-phycocyanin from Galdieria phlegrea: Determinants of thermostability and comparison with a C-phycocyanin in the entire phycobilisome.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6Y3Q
 
 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y4A
 
 | | Crystal structure of the M295I variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-02-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6Y57
 
 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y5Z
 
 | |
6Y60
 
 | | Structure of Human Polyomavirus 12 VP1 in complex with 3'-Sialyllactosamine | | Descriptor: | Capsid protein VP1, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y61
 
 | |
6Y63
 
 | | Structure of Sheep Polyomavirus VP1 in complex with 3'-Sialyllactosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y65
 
 | | Structure of apo Goose Hemorrhagic Polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y66
 
 | | Structure of Goose Hemorrhagic Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y67
 
 | |







