7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N8B
 
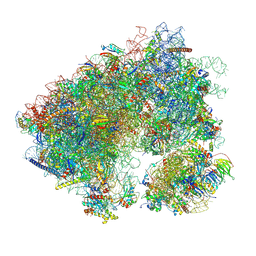 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N9X
 
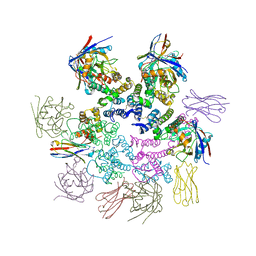 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | Descriptor: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.511 Å) | | Cite: | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7NA6
 
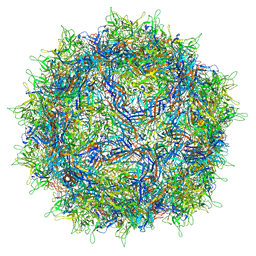 | | Cryo-EM structure of AAV True Type | | Descriptor: | Capsid protein VP1 | | Authors: | Bennett, A.D, McKenna, R. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Comparative structural, biophysical, and receptor binding study of true type and wild type AAV2.
J.Struct.Biol., 213, 2021
|
|
7NAC
 
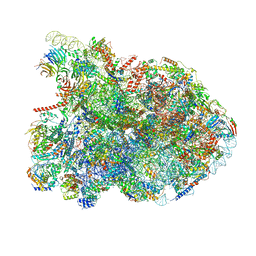 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NB1
 
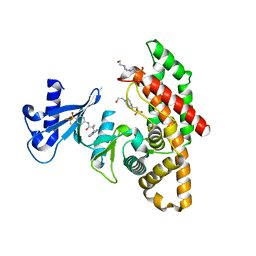 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-aminopurin-9-yl)-~{N}-[4-(trifluoromethylsulfonyl)phenyl]cyclohexane-1-carboxamide, Choline kinase alpha | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7NB2
 
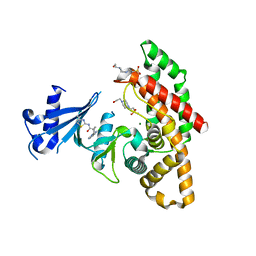 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-azanyl-2-chloranyl-purin-9-yl)-~{N}-(4-methyl-1,3-thiazol-2-yl)cyclohexane-1-carboxamide, Choline kinase alpha, ... | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7NB3
 
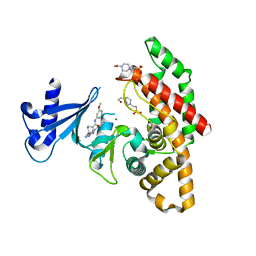 | | Crystal structure of human choline alpha in complex with an inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-(6-azanyl-2-pyridin-4-yl-purin-9-yl)-~{N}-(3-methoxyphenyl)cyclohexane-1-carboxamide, Choline kinase alpha, ... | | Authors: | Casale, E, Fasolini, M. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of unprecedented ATP-competitive choline kinase inhibitors.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7NBC
 
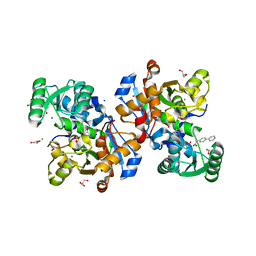 | | Crystal structure of human serine racemase in complex with DSiP fragment Z2856434779, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBD
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z235449082, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBF
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z126932614, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBG
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z52314092, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBH
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z26781964, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBR
 
 | |
7NBS
 
 | |
7NBT
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | Descriptor: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NBU
 
 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7NCX
 
 | | Crystal structure of GH30 (double mutant EE) from Thermothelomyces thermophila. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dimarogona, M, Nikolaivits, E, Topakas, E, Weiss, M, Feiler, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Carbohydrate Polymers, 273, 2021
|
|
7NDQ
 
 | | Gag:02 TCR in complex with HLA-E. | | Descriptor: | Beta-2-microglobulin, Gag6V, HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDT
 
 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDU
 
 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NE9
 
 | |
7NEO
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|







