8A3K
 
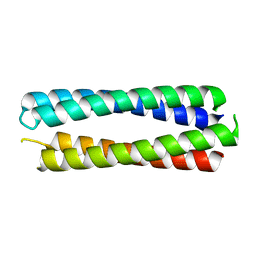 | | X-ray crystal structure of a de novo designed single-chain antiparallel 4-helix coiled-coil bundle, sc-apCC-4 | | Descriptor: | sc-apCC-4 | | Authors: | Albanese, K.I, Mylemans, B, Naudin, E.A, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8A3L
 
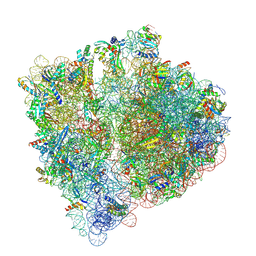 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
8A3W
 
 | |
8A3Y
 
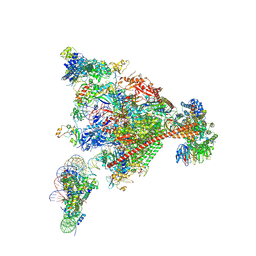 | | Structure of mammalian Pol II-DSIF-SPT6-PAF1-TFIIS-hexasome elongation complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Farnung, L, Ochmann, M, Garg, G, Vos, S.M, Cramer, P. | | Deposit date: | 2022-06-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a backtracked hexasomal intermediate of nucleosome transcription.
Mol.Cell, 82, 2022
|
|
8A47
 
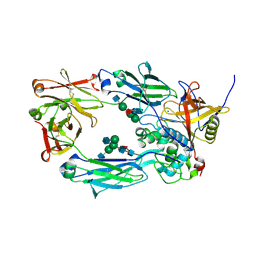 | | IdeS in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG-degrading protease, ... | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
8A4R
 
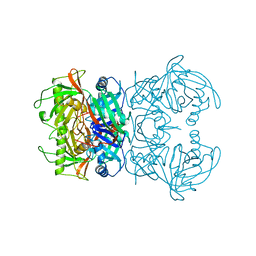 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic orthorhombic data at 3.59 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
8A5N
 
 | |
8A5R
 
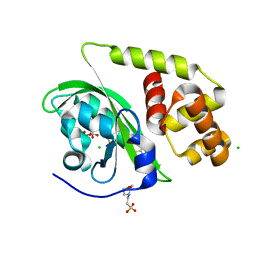 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5S
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5Y
 
 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
8A60
 
 | |
8A64
 
 | | cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1. | | Descriptor: | Endo-beta-N-acetylglucosaminidase F2, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Cifuente, J.O, Du, J.J, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases.
Nat Commun, 14, 2023
|
|
8A94
 
 | | SARS CoV2 Spike in the 2-up state in complex with Fab47. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain Variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A95
 
 | | SARS Cov2 Spike RBD in complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8A97
 
 | | ROOM TEMPERATURE CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) UNDER XENON PRESSURE (30 bar) | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, D(-)-TARTARIC ACID, XENON | | Authors: | Bui, S, Prange, T, Steiner, R.A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8A98
 
 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT | | Descriptor: | 40S ribosomal protein S12, 40S ribosomal protein S14, 40S ribosomal protein S19-like protein, ... | | Authors: | Rajan, K.S, Yonath, A, Bashan, A. | | Deposit date: | 2022-06-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8A9A
 
 | |
8A9E
 
 | | Lysozyme, 9-11 fs FEL pulses as determined by XTCAV | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme | | Authors: | Barends, T, Nass, K, Gorel, A, Schlichting, I. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Microcrystallization methods
To Be Published
|
|
8A9F
 
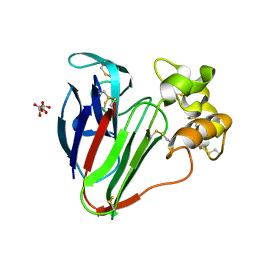 | |
8AA5
 
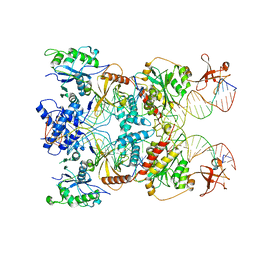 | | Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon) | | Descriptor: | LE_PolyA, LE_Target, RE_PolyA, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Lopez-Mendez, B, Stutzke, L.S, Fuglsang, A, Stella, S, Montoya, G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon.
Nat Commun, 13, 2022
|
|
8AAC
 
 | |
8AAF
 
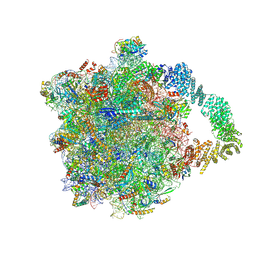 | | Yeast RQC complex in state G | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-01 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8ACM
 
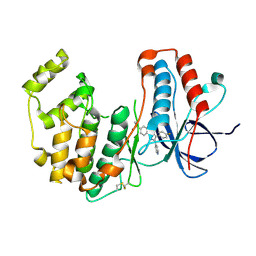 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACO
 
 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACQ
 
 | | S-layer Deinoxanthin-Binding Complex (SDBC), subunit DR_2577 assembled with its SOD DR_0644 | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, DR_0644, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|






