7LWO
 
 | |
7LWP
 
 | |
7LWQ
 
 | |
7LWS
 
 | |
7LWT
 
 | |
7LWU
 
 | |
7LWV
 
 | |
7LWW
 
 | |
7LYK
 
 | |
7LYL
 
 | |
7LYM
 
 | |
7LYN
 
 | |
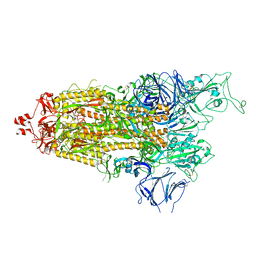
7LYO
 
 | |
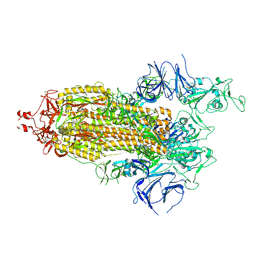
7LYP
 
 | |
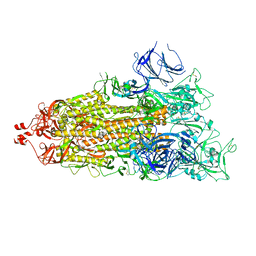
7LYQ
 
 | |
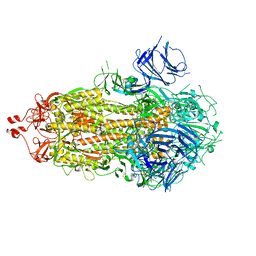
7M0J
 
 | |
7M4R
 
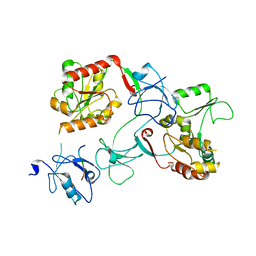 | |
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
7D4G
 
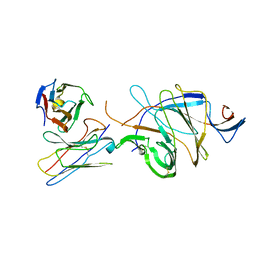 | |
7E3J
 
 | |
7MBG
 
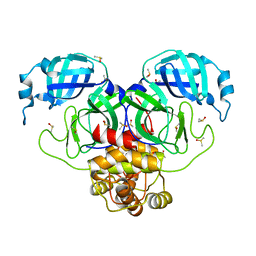 | | SARS-CoV-2 Main protease in orthorhombic space group | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7NF5
 
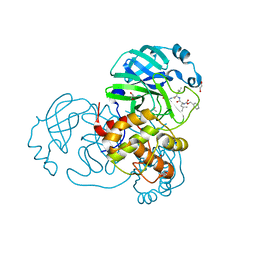 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NG3
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NG6
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NNG
 
 | | Crystal structure of the SARS-CoV-2 helicase in complex with Z2327226104 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|








