9F13
 
 | | Crystal structure of HLA-C*12:02 in complex with KAYNVTQAF (KF9), a 9-mer epitope from SARS-CoV-2 Nucleocapsid (N266-274) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen C alpha chain, Nucleoprotein | | Authors: | Ahn, Y.M, Maddumage, J.C, Chatzileontiadou, D.S.M, Gras, S. | | Deposit date: | 2024-04-18 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Molecular basis of potent antiviral HLA-C-restricted CD8 + T cell response to an immunodominant SARS-CoV-2 nucleocapsid epitope.
Nat Commun, 16, 2025
|
|
9HLJ
 
 | | Crystal structure of GV37-TCR in complex with HLA-C*12:02 with KAYNVTQAF (KF9), a 9-mer epitope from SARS-CoV-2 Nucleocapsid (N266-274) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GV37-TCR alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Chatzileontiadou, D.S.M, Gras, S. | | Deposit date: | 2024-12-05 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis of potent antiviral HLA-C-restricted CD8 + T cell response to an immunodominant SARS-CoV-2 nucleocapsid epitope.
Nat Commun, 16, 2025
|
|
9JRC
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain complexed with squirrel ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lan, J, Nan, X. | | Deposit date: | 2024-09-29 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Cross-species recognition of squirrel ACE2 by the receptor binding domains of SARS-CoV-2, RaTG13, PCoV-GD and PCoV-GX.
Structure, 33, 2025
|
|
9KUD
 
 | | Crystal structure of SARS-CoV-2 JN.1 variant RBD complexed with squirrel ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Lan, J, Wang, C.H. | | Deposit date: | 2024-12-03 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Cross-species recognition of squirrel ACE2 by the receptor binding domains of SARS-CoV-2, RaTG13, PCoV-GD and PCoV-GX.
Structure, 33, 2025
|
|
9M2V
 
 | |
9J4S
 
 | |
9J4T
 
 | |
9J4U
 
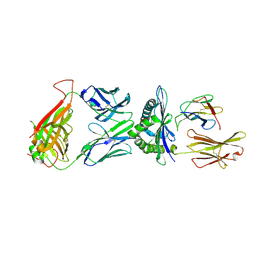 | |
9J4V
 
 | |
9QWI
 
 | |
9RU5
 
 | | Cryo-EM structure of TCRpub/pMHC | | Descriptor: | MHC class I antigen, ORF3a protein, TCRpub alpha chain, ... | | Authors: | Akil, C, Zhu, Y, Hardenbrook, N, Zhang, P. | | Deposit date: | 2025-07-03 | | Release date: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structure of TCRpub/pMHC
To Be Published
|
|
9ECZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with human neutralizing antibody WRAIR-2008 (focused refinement of NTD and WRAIR-2008) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jensen, J.L, Thomas, P.V, Joyce, M.G. | | Deposit date: | 2024-11-15 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | First-generation N-terminal domain supersite public antibodies retain activity against Omicron-derived lineages and protect mice against Omicron BA.5 challenge.
Mbio, 16, 2025
|
|
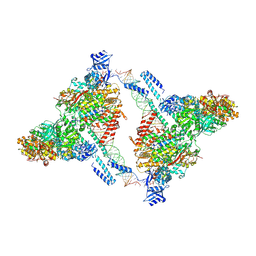
9IMK
 
 | |
9IMM
 
 | |
9KGJ
 
 | |
9KGQ
 
 | |
9KGR
 
 | |
9KVD
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with triple-nAb 3G5, 4H5 and 4C11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, The heavy chain of 3G5, ... | | Authors: | Sun, H, Jiang, Y, Wang, S, Zheng, Z, Li, S, Zheng, Q. | | Deposit date: | 2024-12-05 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with triple-nAb 3G5, 4H5 and 4C11
To Be Published
|
|
9KVE
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with triple-nAb 4H1, 4A5 and 4C1 | | Descriptor: | Spike protein S1, The heavy chain of 4A5, The heavy chain of 4C1, ... | | Authors: | Sun, H, Jiang, Y, Wang, S, Zheng, Z, Li, S, Zheng, Q. | | Deposit date: | 2024-12-05 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with triple-nAb 4H1, 4A5 and 4C1
To Be Published
|
|
9KVF
 
 | | Cryo-EM structure of SARS-CoV-2 EG.1 spike protein in complex with triple-nAb 4A5, 4C1 and 2E10 | | Descriptor: | 2E10 heavy chain, 2E10 light chain, 4A5 heavy chain, ... | | Authors: | Sun, H, Jiang, Y, Wang, S, Zheng, Z, Li, S, Zheng, Q. | | Deposit date: | 2024-12-05 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 EG.1 spike protein in complex with triple-nAb 4A5, 4C1 and 2E10
To Be Published
|
|
9MI3
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with neutralizing human antibody WRAIR-2008 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, WRAIR-2008 antibody Fab heavy chain, ... | | Authors: | Jensen, J.L, Thomas, P.V, Joyce, M.G. | | Deposit date: | 2024-12-12 | | Release date: | 2025-08-20 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | First-generation N-terminal domain supersite public antibodies retain activity against Omicron-derived lineages and protect mice against Omicron BA.5 challenge.
Mbio, 16, 2025
|
|
9N3M
 
 | | SARS-CoV-2 Mpro L50F/E166A/L167F triple mutant bound to inhibitor | | Descriptor: | (1S,2S,4S)-2-{[3-cyclopropyl-N-(4-methoxy-1H-indole-2-carbonyl)-L-alanyl]amino}-1-hydroxy-4-methyl-5-(methylamino)-5-oxopentane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lu, J, Chen, P, Demmon, S, Fischer, C, Vederas, J.C, Lemieux, M.J. | | Deposit date: | 2025-01-31 | | Release date: | 2025-08-20 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the nirmatrelvir-resistant SARS-CoV-2 M pro L50F/E166A/L167F triple mutant-inhibitor-complex reveal strategies for next generation coronaviral inhibitor design.
Rsc Med Chem, 2025
|
|
9UCN
 
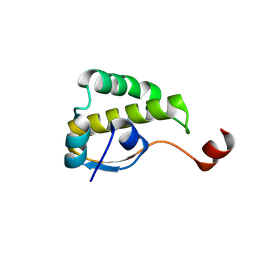 | | Monomer of SARS-CoV-2 nsp4CTD | | Descriptor: | Non-structural protein 4 | | Authors: | Meng, L, Pei, K, Tang, C. | | Deposit date: | 2025-04-04 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Dynamics of SARS-CoV-2 NSP4 C-terminal Domain and Implications for Viral Processing.
J.Mol.Biol., 437, 2025
|
|
9ATQ
 
 | | XBB.1.5 spike/Nanosota-8 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of XBB.1.5 spike and Nanosota-8 complex
To Be Published
|
|
9ATR
 
 | | local refinement of XBB.1.5 spike/Nanosota-8 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-8, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of XBB.1.5 spike and Nanosota-3C complex
To Be Published
|
|








