9LDS
 
 | |
9LYO
 
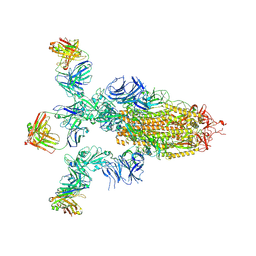 | | Alpha SARS-CoV-2 spike protein in complex with REGN10987 Fab homologue. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, REGN10987 Fab homologue (Heavy chain), ... | | Authors: | Kocharovskaya, M.V, Pichkur, E.B, Shenkarev, Z.O, Lyukmanova, E.N. | | Deposit date: | 2025-02-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and dynamics of Alpha B.1.1.7 SARS-CoV-2 S-protein in complex with Fab of neutralizing antibody REGN10987.
Biochem.Biophys.Res.Commun., 755, 2025
|
|
9LYP
 
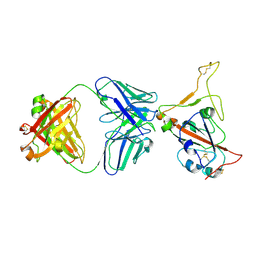 | | Alpha SARS-CoV-2 spike protein RBD-down in complex with REGN10987 Fab homologue (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, REGN10987 Fab homologue (Heavy chain), REGN10987 Fab homologue (Light chain), ... | | Authors: | Kocharovskaya, M.V, Pichkur, E.B, Shenkarev, Z.O, Lyukmanova, E.N. | | Deposit date: | 2025-02-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of Alpha B.1.1.7 SARS-CoV-2 S-protein in complex with Fab of neutralizing antibody REGN10987.
Biochem.Biophys.Res.Commun., 755, 2025
|
|
8YWY
 
 | | Crystal structure of SARS-Cov-2 main protease E166N mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YWZ
 
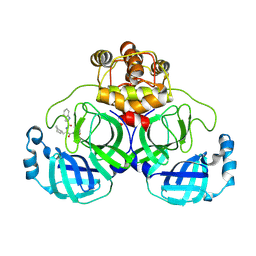 | | Crystal structure of SARS-Cov-2 main protease H163A mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zou, X.F, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9INL
 
 | | Crystal structure of SARS-Cov-2 main protease E166R mutant in complex with Bofutrelvir | | Descriptor: | Replicase polyprotein 1a, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2024-07-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9LUF
 
 | | Crystal structure of SARS-Cov-2 main protease S144A mutant in complex with Bofutrelvir | | Descriptor: | ORF1a polyprotein, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2025-02-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9LUG
 
 | | Crystal structure of SARS-Cov-2 main protease E166V mutant in complex with Bofutrelvir | | Descriptor: | ORF1a polyprotein, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2025-02-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9B4X
 
 | |
9B9U
 
 | |
9BD9
 
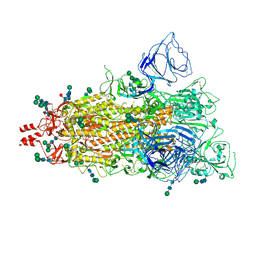 | | SARS CoV-2 full-length WT spike protein, 1RBD-up conformation (SPIKE-WT) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-04-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of SARS CoV-2 full-length spike protein, 1RBD-up conformation
To Be Published
|
|
9DHY
 
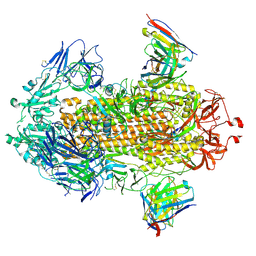 | | Structure of SARS-CoV-2 spike in complex with antibody Fab COVIC-154 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab COVIC-154 Heavy Chain, ... | | Authors: | Yu, X, Saphire, E.O. | | Deposit date: | 2024-09-04 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A global collaboration for systematic analysis of broad-ranging antibodies against the SARS-CoV-2 spike protein.
Cell Rep, 44, 2025
|
|
8Z2E
 
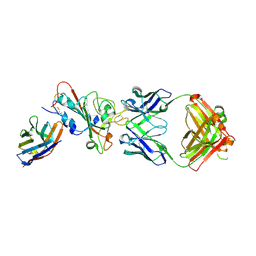 | | Crystal structure of nanobody Tnb04-1 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Chen, J, Dong, H.D. | | Deposit date: | 2024-04-12 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of nanobody Tnb04-1 with antibody 1F11 fab and SARS-CoV-2 RBD
To Be Published
|
|
9BC1
 
 | |
9KN1
 
 | |
8Z9L
 
 | |
8ZBF
 
 | |
9DU2
 
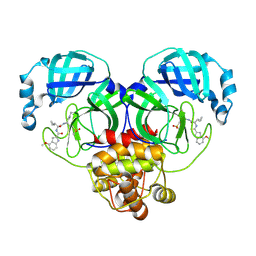 | | SARS-CoV-2 Mpro in complex with compound 7 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-{[(1Z,2S)-1-imino-4-(methanesulfonyl)butan-2-yl]amino}-1-oxopropan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bigelow, L, Tang, H, Boguslaw, N, Duda, D.M. | | Deposit date: | 2024-10-02 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inhibition of dimeric SARS-CoV-2 Mpro displays positive cooperativity and a mixture of covalent and non-covalent binding.
Iscience, 28, 2025
|
|
9FM4
 
 | | Dynamic structure of the apical stem loop of the stem loop 2 motif (s2m) from SCoV-2 Delta variant | | Descriptor: | RNA (25-MER) | | Authors: | Wirtz Martin, M.A, Matzel, T, Makowski, J, Kensinger, A, Herr, A, Wacker, A, Richter, C, Jonker, H.R.A, Evanseck, J, Schwalbe, H. | | Deposit date: | 2024-06-05 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural heterogeneity and dynamics in the apical stem loop of s2m from SARS-CoV-2 Delta by an integrative NMR spectroscopy and MD simulation approach.
Nucleic Acids Res., 53, 2025
|
|
8X1X
 
 | | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Choudhary, S, Verma, S, Kumar, P, Tomar, S. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid
To Be Published
|
|
9BLF
 
 | | SARS-CoV-2 core polymerase complex inhibited by araCTP | | Descriptor: | 4-amino-1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Anderson, T.K, Kirchdeorfer, R.N. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of the SARS-CoV-2 core-RTC bound to RNA, araCMP, and araCTP
To Be Published
|
|
9C2H
 
 | | SARS-CoV-2 Nucleocapsid Dimerization Domain bound to Fab-NP1E9 and Fab-NP3B4 | | Descriptor: | Antibody Fab NP1-E9 Heavy Chain (variable region), Antibody Fab NP1-E9 Light Chain (variable region), Antibody Fab NP3-B4 Heavy Chain (variable region), ... | | Authors: | Landeras-Bueno, S, Hariharan, C, Diaz Avalos, R, Ollmann Saphire, E. | | Deposit date: | 2024-05-31 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural stabilization of the intrinsically disordered SARS-CoV-2 N by binding to RNA sequences engineered from the viral genome fragment.
Nat Commun, 16, 2025
|
|
9F7P
 
 | |
9F7R
 
 | |
9F7S
 
 | |








