8TQU
 
 | |
9FJK
 
 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Broadly potent spike-specific human monoclonal antibodies inhibit SARS-CoV-2 Omicron sub-lineages.
Commun Biol, 7, 2024
|
|
8UPX
 
 | | Omicron-S-MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 omicron spike with MERS RBD | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-10-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.
J.Virol., 98, 2024
|
|
8YE4
 
 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | Descriptor: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
8ZHE
 
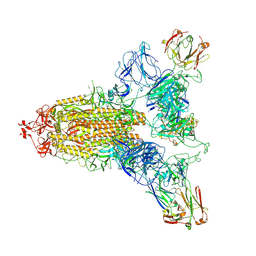 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHG
 
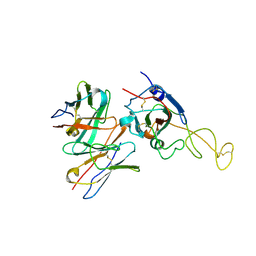 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, focused refinement of RBD-Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, Light chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHH
 
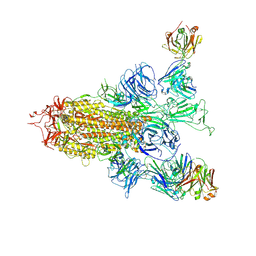 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (5.55 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHM
 
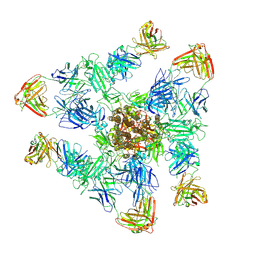 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHO
 
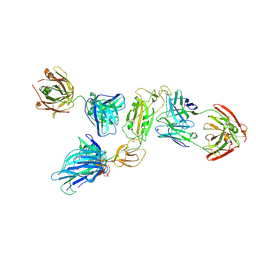 | | SARS-CoV-2 S1 in complex with H18 and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, Heavy chain of R1-32 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZV9
 
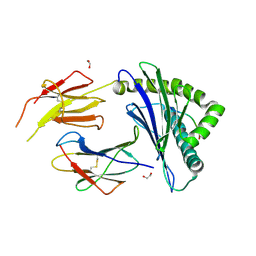 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
9F9Y
 
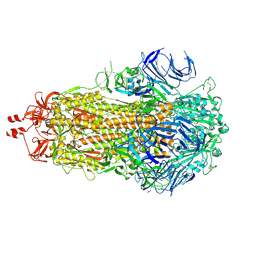 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
8UG9
 
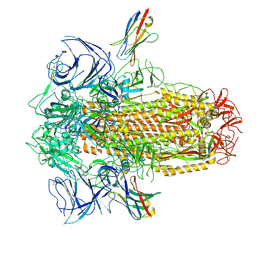 | | XBB.1.5 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
8WTI
 
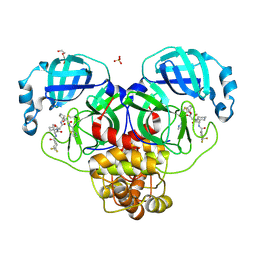 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
9FEH
 
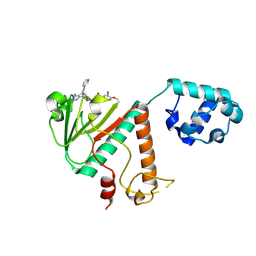 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | Descriptor: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | Authors: | Zilecka, E, Klima, M, Boura, E. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
8KEH
 
 | |
8Y4C
 
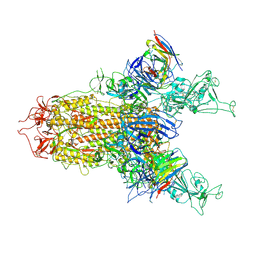 | | BA.2.86 S-trimer in complex with Nab XGv280 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XGv280 Heavy chain, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
8ZER
 
 | | Crystal structure of the complex of Wuhan SARS-CoV-2 RBD (319-541) with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, Spike protein S1, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
8ZES
 
 | | Crystal structure of the Wuhan SARS-CoV-2 RBD (333-541) complexed with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
9EO6
 
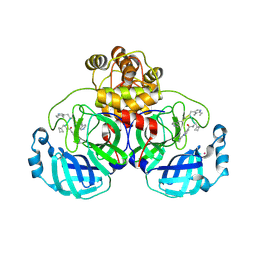 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOX
 
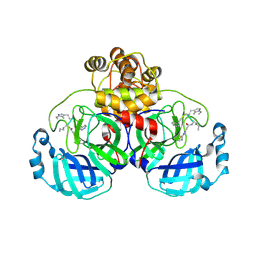 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
8Y4A
 
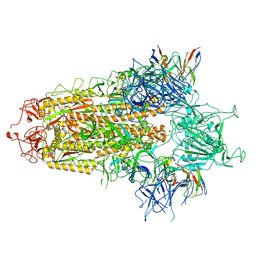 | | BA.2.86 S-trimer in complex with Nab XG2v046 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
8U3O
 
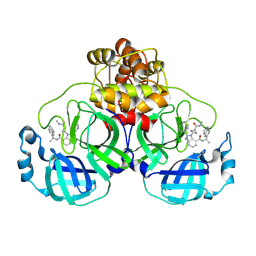 | | SARS-CoV-2 Main Protease A173V in complex with CDD-1819 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, ORF1a polyprotein | | Authors: | Nnabuife, C, Palzkill, T. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | SARS-CoV-2 Main Protease A173V in complex with CDD-1819
To Be Published
|
|
8ZBQ
 
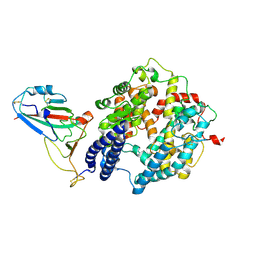 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
9ATO
 
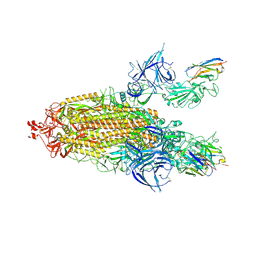 | | XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3C, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided in vitro evolution of nanobodies targeting new viral variants.
Plos Pathog., 20, 2024
|
|








