8JYS
 
 | |
8UIR
 
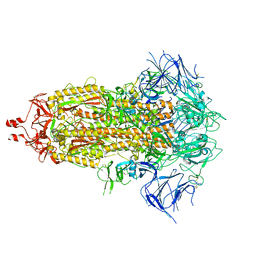 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8V0L
 
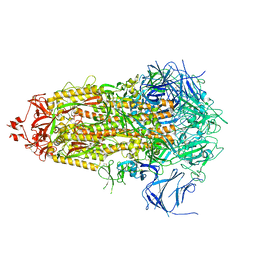 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer 1 (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8XZD
 
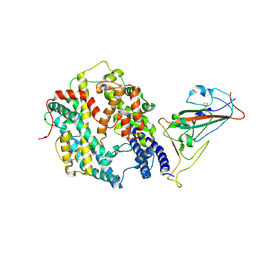 | | The structure of fox ACE2 and Omicron BF.7 RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
9AYW
 
 | |
9FGS
 
 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv41N (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single Chain fragment scFv41N, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv41N in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9FGT
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain fragment scFv76, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of scFv76 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
9FGU
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
8C0Y
 
 | |
8QH0
 
 | | Crystal structure of the SARS-CoV-2 RBD with the antibody Cv2.3194 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cv2.3194 Heavy chain, GLYCEROL, ... | | Authors: | Fernandez, I, Rey, F.A. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Broad sarbecovirus neutralization by combined memory B cell antibodies to ancestral SARS-CoV-2.
Iscience, 27, 2024
|
|
8QH1
 
 | |
8UKD
 
 | |
8UKF
 
 | | SARS-CoV-2 Omicron-EG.5 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-EG.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8XBF
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8XXW
 
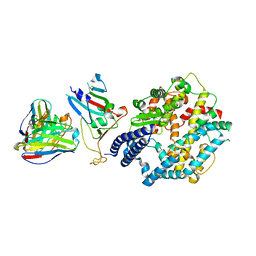 | | Fab M2-7 complexed with SARS-Cov2 RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, M2-7-Heavy chain, M2-7-Light chain, ... | | Authors: | Liu, C, Xie, Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mosaic RBD nanoparticle elicits immunodominant antibody responses across sarbecoviruses.
Cell Rep, 43, 2024
|
|
8YB5
 
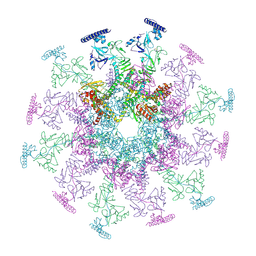 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
9EOW
 
 | |
8T7Y
 
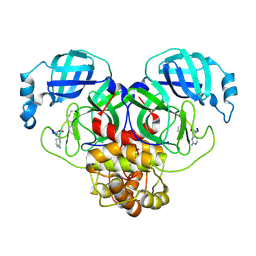 | |
8WHZ
 
 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4Z
 
 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8OYT
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8WHU
 
 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WP8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-09 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8X5Q
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|








