7R15
 
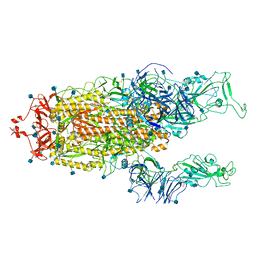 | | Alpha Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R1A
 
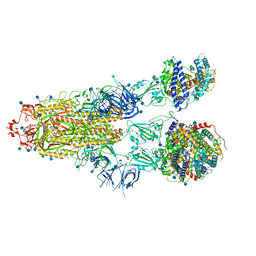 | | Furin Cleaved Alpha Variant SARS-CoV-2 Spike in complex with 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7THK
 
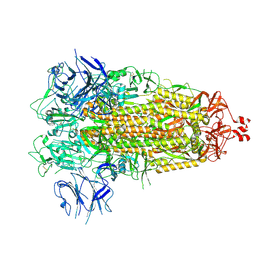 | |
7TZJ
 
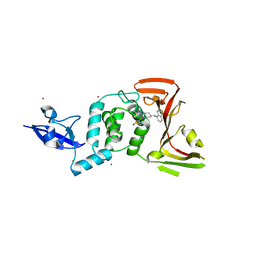 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
7VVT
 
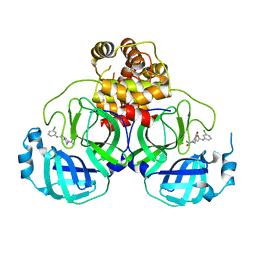 | | SARS-CoV-2 3CL protease (3CLpro) in complex with a covalent inhibitor | | Descriptor: | 3C-like proteinase, N-(3-chlorophenyl)-2-[(2R)-1-ethanoyl-3-oxidanylidene-piperazin-2-yl]ethanamide | | Authors: | Su, H, Nie, T, Li, M, Xu, Y. | | Deposit date: | 2021-11-08 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | In silico screening-based discovery of novel covalent inhibitors of the SARS-CoV-2 3CL protease.
Eur.J.Med.Chem., 231, 2022
|
|
7VYR
 
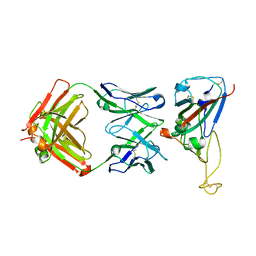 | |
7YWR
 
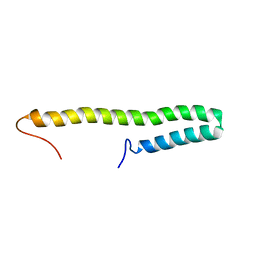 | |
7E9O
 
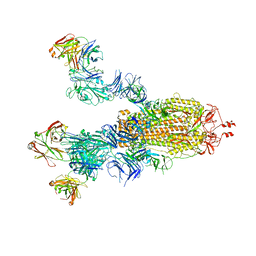 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7Q1Z
 
 | | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Sulbaran, G, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.
Cell Rep Med, 3, 2022
|
|
7R0Z
 
 | | Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R11
 
 | | Dissociated S1 domain of Beta Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R12
 
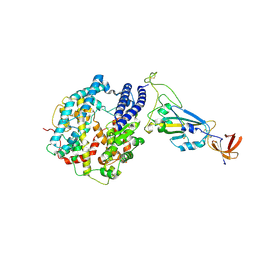 | | Dissociated S1 domain of Mink Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R13
 
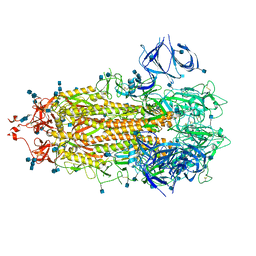 | | Alpha Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R14
 
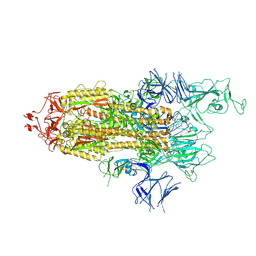 | | Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R16
 
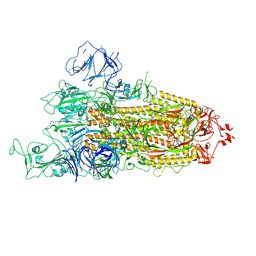 | | Beta Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R17
 
 | | Beta Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R18
 
 | | Mink Variant SARS-CoV-2 Spike in Closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R19
 
 | | Mink Variant SARS-CoV-2 Spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R1B
 
 | | Mink Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7R2V
 
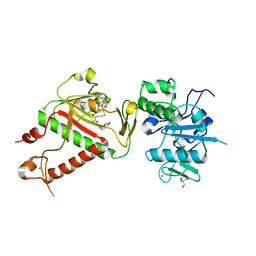 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
7S5P
 
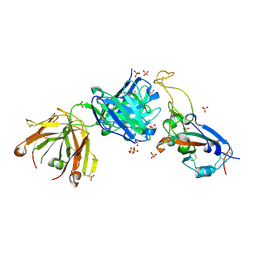 | |








