9FCM
 
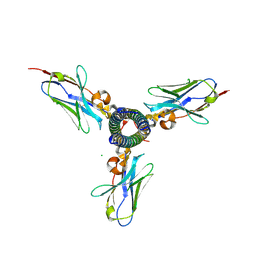 | | Single-domain antibody binding the SARS-COV2 S2 | | Descriptor: | CHLORIDE ION, Single-domain antibody R3DC23, Spike protein S2' | | Authors: | Vann Molle, I, Remaut, H. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ultrapotent SARS coronavirus-neutralizing single-domain antibodies that clamp the spike at its base.
Nat Commun, 16, 2025
|
|
9UJ2
 
 | | 14-3-3 zeta chimera with the S202R peptide of SARS-CoV-2 N (residues 200-213) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta,Peptide from Nucleoprotein, DI(HYDROXYETHYL)ETHER | | Authors: | Boyko, K.M, Matyuta, I.O, Minyaev, M.E, Perfilova, K.V, Sluchanko, N.N. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Biochem.Biophys.Res.Commun., 767, 2025
|
|
9ELE
 
 | |
9ELF
 
 | |
9ELG
 
 | |
9ELH
 
 | |
9ELI
 
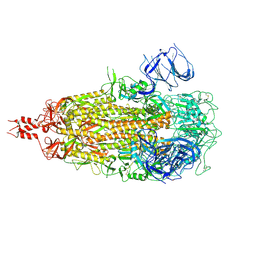 | |
9ELN
 
 | |
9ELO
 
 | |
9ELP
 
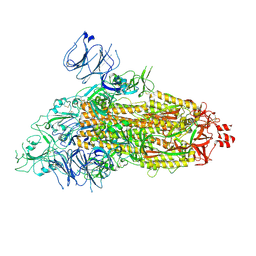 | |
9ELQ
 
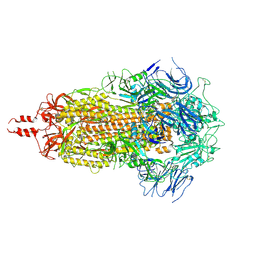 | |
9HH5
 
 | | Crystal Structure of nsp15 Endoribonuclease from SARS CoV-2 in Complex with Sepantronium (YM-155) | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nsp15 - Uridylate-specific endoribonuclease, ... | | Authors: | Chatziefthymiou, S.D, Kolbe, M, Hakanpaeae, J, Labahn, J, Windshuegel, B. | | Deposit date: | 2024-11-21 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification, validation, and characterization of approved and investigational drugs interfering with the SARS-CoV-2 endoribonuclease Nsp15.
Protein Sci., 34, 2025
|
|
9HH6
 
 | | Crystal Structure of Nsp15 Endoribonuclease from SARS CoV-2 in Complex with TAS-103 | | Descriptor: | 1,2-ETHANEDIOL, 6-[2-(dimethylamino)ethylamino]-3-oxidanyl-indeno[2,1-c]quinolin-7-one, ORF1ab polyprotein, ... | | Authors: | Chatziefthymiou, S.D, Kolbe, M, Hakanpaeae, J, Labahn, J, Windshuegel, B. | | Deposit date: | 2024-11-21 | | Release date: | 2025-05-28 | | Method: | SOLUTION SCATTERING (2 Å), X-RAY DIFFRACTION | | Cite: | Identification, validation, and characterization of approved and investigational drugs interfering with the SARS-CoV-2 endoribonuclease Nsp15.
Protein Sci., 34, 2025
|
|
9IZB
 
 | |
9ELJ
 
 | |
9ELK
 
 | |
9ELL
 
 | |
9ELM
 
 | |
9ML4
 
 | |
9ML5
 
 | |
9ML6
 
 | |
9ML7
 
 | |
9ML8
 
 | |
9ML9
 
 | |
9MRU
 
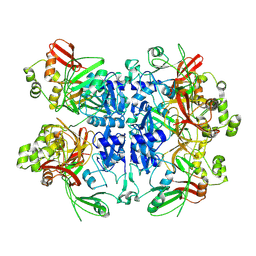 | | Structural Asymmetry in SARS-CoV-2 Nsp15 Hexamer Important for Catalytic Activity | | Descriptor: | Uridylate-specific endoribonuclease nsp15 | | Authors: | Ketawala, G.K, Sonowal, M, Schrag, L, Fromme, R, Botha, S, Fromme, P. | | Deposit date: | 2025-01-08 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Asymmetry in SARS-CoV-2 Nsp15 Hexamer Important for Catalytic Activity
To Be Published
|
|








