8YLS
 
 | | Structure of SARS-CoV-2 Mpro in complex with its degrader | | Descriptor: | (4-methoxyphenyl)methyl ~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Feng, Y, Li, W, Cheng, S.H, Li, X.B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Development of novel antivrial agents that induce the degradation of the main protease of human-infecting coronaviruses.
Eur.J.Med.Chem., 275, 2024
|
|
8ZER
 
 | | Crystal structure of the complex of Wuhan SARS-CoV-2 RBD (319-541) with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, Spike protein S1, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
8ZES
 
 | | Crystal structure of the Wuhan SARS-CoV-2 RBD (333-541) complexed with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
9EO6
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOR
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL12. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL12, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9EOX
 
 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
8AJ1
 
 | | SARS-CoV-2 Mpro in Complex with RK-107 | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(phenylcarbamoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2024-09-11 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8Y4A
 
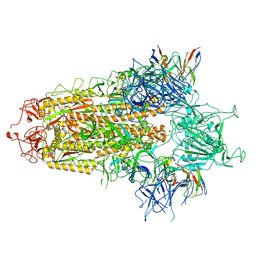 | | BA.2.86 S-trimer in complex with Nab XG2v046 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
8U3O
 
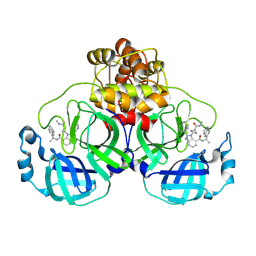 | | SARS-CoV-2 Main Protease A173V in complex with CDD-1819 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, ORF1a polyprotein | | Authors: | Nnabuife, C, Palzkill, T. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | SARS-CoV-2 Main Protease A173V in complex with CDD-1819
To Be Published
|
|
8ZBQ
 
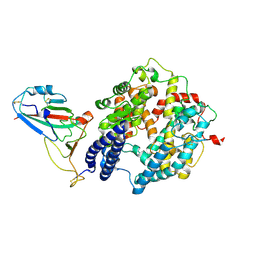 | | Local map of Omicron Subvariant JN.1 RBD with ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Yan, R.H, Yang, H.N. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
9ATO
 
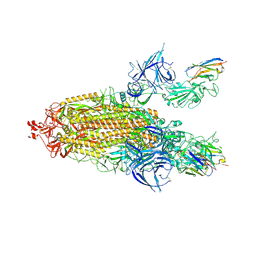 | | XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3C, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided in vitro evolution of nanobodies targeting new viral variants.
Plos Pathog., 20, 2024
|
|
9ATP
 
 | | local refinement of XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | Nanosota-3C, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-guided in vitro evolution of nanobodies targeting new viral variants.
Plos Pathog., 20, 2024
|
|
8U9H
 
 | |
8U9K
 
 | |
8U9M
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI95 | | Descriptor: | 3C-like proteinase nsp5, bis(4-fluorophenyl)methyl (1R,2S,5R)-2-({(2R)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-3-carboxylate | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
8U9N
 
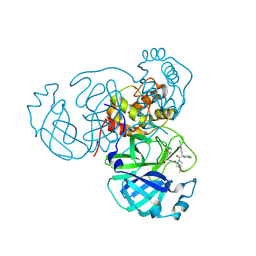 | |
8U9T
 
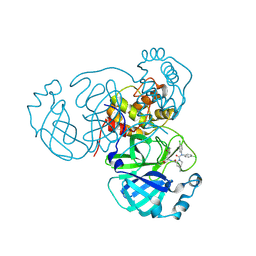 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI97 | | Descriptor: | (1R,2S,5S)-N~3~,N~3~-bis(4-chlorophenyl)-N~2~-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
8U9U
 
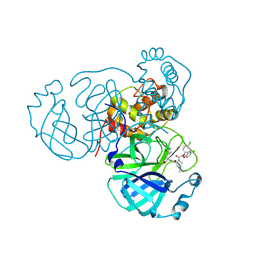 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI98 | | Descriptor: | (1R,2S,5S)-3-[bis(4-chlorophenoxy)acetyl]-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
8U9V
 
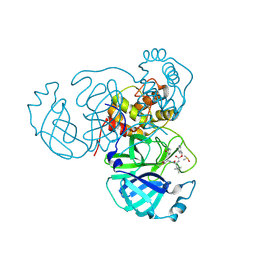 | |
8U9W
 
 | |
8WFH
 
 | | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1 | | Descriptor: | D1 scFv, Spike protein S1 | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1
To Be Published
|
|
8WFM
 
 | | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11 | | Descriptor: | Spike protein S1, T11 scFv | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11
To Be Published
|
|
9DN4
 
 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic studies of small molecule ligands selective to RNA single G bulges.
Nucleic Acids Res., 53, 2025
|
|
9GNY
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and Caffeine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-04 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
to be published
|
|
9GRP
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and beta-chloroethyl theophylline | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|








