7TF8
 
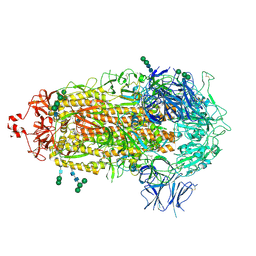 | |
7THE
 
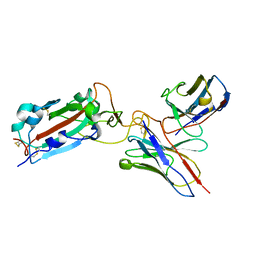 | | Structure of RBD directed antibody DH1042 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interface | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 Fab Heavy Chain, DH1042 Fab Light Chain, ... | | 著者 | May, A.J, Manne, K, Acharya, P. | | 登録日 | 2022-01-10 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
7THT
 
 | | CryoEM structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1042 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1042 heavy chain, ... | | 著者 | Manne, K, May, A, Acharya, P. | | 登録日 | 2022-01-12 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structural diversity of the SARS-CoV-2 Omicron spike.
Mol.Cell, 82, 2022
|
|
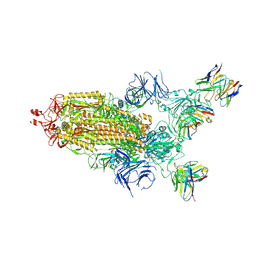
7TNW
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-22 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TOW
 
 | |
7TVX
 
 | |
7AU4
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 3 | | 分子名称: | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Talibov, V.O. | | 登録日 | 2020-11-02 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2J
 
 | |
7B2U
 
 | |
7B5Z
 
 | |
7B77
 
 | |
7BIJ
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 13 | | 分子名称: | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Talibov, V.O. | | 登録日 | 2021-01-12 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7E6K
 
 | | Viral protease | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(2-phenoxyethyl)methanethioamide, ... | | 著者 | Zhao, Y, Zhang, Q, Yang, H.T, Rao, Z.H. | | 登録日 | 2021-02-22 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Viral protease
To be published
|
|
7NBT
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | 分子名称: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Talibov, V.O. | | 登録日 | 2021-01-27 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NEO
 
 | |
7O46
 
 | |
7Q0A
 
 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7QBB
 
 | |
7TL1
 
 | |
7TL9
 
 | |
7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | 分子名称: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TWF
 
 | |
7TWG
 
 | |








