8Z7G
 
 | |
9CDK
 
 | | SARS-CoV-2 Mpro A173V mutant in complex with small molecule inhibitor Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Tang, S, Kenneson, J.R, Anderson, K.S. | | Deposit date: | 2024-06-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9CDL
 
 | | SARS-CoV-2 Mpro E166V/L50F double mutant in complex with small molecule inhibitor Mpro61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Tang, S, Kenneson, J.R, Huynh, K, Anderson, K.S. | | Deposit date: | 2024-06-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9G9C
 
 | |
9G9D
 
 | |
8XSD
 
 | | BA.5 Spike complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike glycoprotein | | Authors: | Feng, L.L. | | Deposit date: | 2024-01-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
8Z6S
 
 | |
8Z6W
 
 | |
8Z86
 
 | | BA.5 RBD in complex with CR9 | | Descriptor: | CR9 heavy chain, CR9 light chain, Spike protein S1 | | Authors: | Feng, L.L. | | Deposit date: | 2024-04-21 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A broadly neutralizing antibody against the SARS-CoV-2 Omicron sub-variants BA.1, BA.2, BA.2.12.1, BA.4, and BA.5.
Signal Transduct Target Ther, 10, 2025
|
|
9BNU
 
 | | Crystal Structure of T190I SARS-CoV-2 Main Protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BNW
 
 | |
9BO7
 
 | | Crystal Structure of L50F SARS-CoV-2 Main Protease in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Papini, C, Kenneson, J, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9BO8
 
 | | Crystal Structure of T21I SARS-CoV-2 Main Protease in Complex with GC376 | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Papini, C, Anderson, K.S. | | Deposit date: | 2024-05-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Exploring Possible Drug-Resistant Variants of SARS-CoV-2 Main Protease (M pro ) with Noncovalent Preclinical Candidate, Mpro61.
Acs Bio Med Chem Au, 5, 2025
|
|
9EX8
 
 | |
9EYA
 
 | |
9EZ4
 
 | | Complex of a mutant of the SARS-CoV-2 main protease Mpro with the nsp5/6 substrate peptide. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | Authors: | Battistutta, R, Fornasier, E, Giachin, G. | | Deposit date: | 2024-04-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allostery in homodimeric SARS-CoV-2 main protease.
Commun Biol, 7, 2024
|
|
9EZ6
 
 | | Complex of a mutant of the SARS-CoV-2 main protease Mpro with the nsp14/15 substrate peptide. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Battistutta, R, Fornasier, E, Giachin, G. | | Deposit date: | 2024-04-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Allostery in homodimeric SARS-CoV-2 main protease.
Commun Biol, 7, 2024
|
|
9J19
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 3C-like proteinase nsp5 | | Authors: | Singh, A, Jangid, K, Dhaka, P, Tomar, S, Kumar, P. | | Deposit date: | 2024-08-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into the Main Protease (Mpro) Dimer Interface Destabilization Inhibitor: Unveiling New Therapeutic Avenues against SARS-CoV-2.
Biochemistry, 64, 2025
|
|
9LAE
 
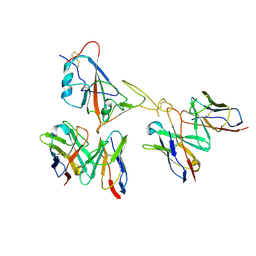 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies 9G11 and 3E2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 3E2, Heavy chain of 9G11, ... | | Authors: | Jiang, Y, Sun, H, Zheng, Q, Li, S. | | Deposit date: | 2025-01-02 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural insight into broadening SARS-CoV-2 neutralization by an antibody cocktail harbouring both NTD and RBD potent antibodies.
Emerg Microbes Infect, 13, 2024
|
|
8VYA
 
 | | SARS-CoV-2 Omicron Variant Spike Glycoprotein Fusion Core (Q954H) | | Descriptor: | SARS-CoV-2 Omicron variant spike glycoprotein C-terminal heptad repeat domain, SARS-CoV-2 Omicron variant spike glycoprotein N-terminal heptad repeat domain (Q954H) | | Authors: | Outlaw, V.K, Apurba, R, Vithanage, N. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | SARS-CoV-2 Omicron Variant Spike Glycoprotein Mutation Q954H Enhances Fusion Core Stability.
Acs Chem.Biol., 20, 2025
|
|
8Z4W
 
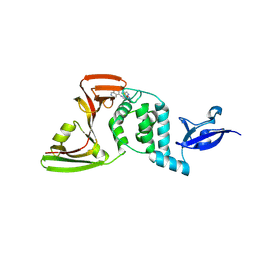 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | 1-[4-[[[4-(isoquinolin-5-ylamino)-6-(methylamino)-1,3,5-triazin-2-yl]amino]methyl]piperidin-1-yl]ethanone, GLYCEROL, Non-structural protein 3, ... | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Nonstructural Protein Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
9B0Y
 
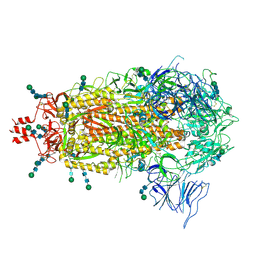 | | SARS CoV-2 Spike protein Ectodomain with internal tag, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, S, Hasan, S.S. | | Deposit date: | 2024-03-12 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Production and cryo-electron microscopy structure of an internally tagged SARS-CoV-2 spike ecto-domain construct.
J Struct Biol X, 11, 2025
|
|
9BJ2
 
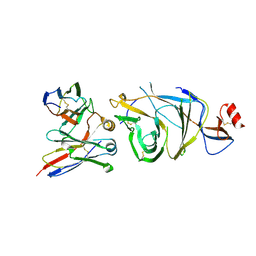 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1533 (local refinement of NTD and C1533) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1533 Heavy Chain, C1533 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
9BJ3
 
 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1596 Fab Heavy Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
9BJ4
 
 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C952 Heavy Chain, C952 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|








