[English] 日本語
 Yorodumi
Yorodumi- PDB-7suc: XFEL Serial Crystallography Reveals the Room Temperature Structur... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7suc | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
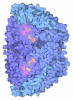
| Title | XFEL Serial Crystallography Reveals the Room Temperature Structure of Methyl-Coenzyme M Reductase | ||||||||||||||||||||||||||||||
 Components Components | (Methyl-coenzyme M reductase I subunit ...) x 3 | ||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSFERASE / Methyl-Coenzyme M Reductase (MCR) / Methanogens / METAL BINDING PROTEIN | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme-B sulfoethylthiotransferase / coenzyme-B sulfoethylthiotransferase activity / methanogenesis / metal ion binding / cytoplasm Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |   Methanothermobacter marburgensis str. Marburg (archaea) Methanothermobacter marburgensis str. Marburg (archaea) | ||||||||||||||||||||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FREE ELECTRON LASER / FREE ELECTRON LASER /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Ohmer, C.J. / Dasgupta, M. | ||||||||||||||||||||||||||||||
| Funding support |  United States, 9items United States, 9items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J.Inorg.Biochem. / Year: 2022 Journal: J.Inorg.Biochem. / Year: 2022Title: XFEL serial crystallography reveals the room temperature structure of methyl-coenzyme M reductase. Authors: Ohmer, C.J. / Dasgupta, M. / Patwardhan, A. / Bogacz, I. / Kaminsky, C. / Doyle, M.D. / Chen, P.Y. / Keable, S.M. / Makita, H. / Simon, P.S. / Massad, R. / Fransson, T. / Chatterjee, R. / ...Authors: Ohmer, C.J. / Dasgupta, M. / Patwardhan, A. / Bogacz, I. / Kaminsky, C. / Doyle, M.D. / Chen, P.Y. / Keable, S.M. / Makita, H. / Simon, P.S. / Massad, R. / Fransson, T. / Chatterjee, R. / Bhowmick, A. / Paley, D.W. / Moriarty, N.W. / Brewster, A.S. / Gee, L.B. / Alonso-Mori, R. / Moss, F. / Fuller, F.D. / Batyuk, A. / Sauter, N.K. / Bergmann, U. / Drennan, C.L. / Yachandra, V.K. / Yano, J. / Kern, J.F. / Ragsdale, S.W. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7suc.cif.gz 7suc.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7suc.ent.gz pdb7suc.ent.gz | 765.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7suc.json.gz 7suc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/su/7suc https://data.pdbj.org/pub/pdb/validation_reports/su/7suc ftp://data.pdbj.org/pub/pdb/validation_reports/su/7suc ftp://data.pdbj.org/pub/pdb/validation_reports/su/7suc | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7sxmC  3m1vS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-Methyl-coenzyme M reductase I subunit ... , 3 types, 6 molecules AaBbCc
| #1: Protein | Mass: 60377.273 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Methanothermobacter marburgensis str. Marburg (archaea) Methanothermobacter marburgensis str. Marburg (archaea)Strain: ATCC BAA-927 / DSM 2133 / JCM 14651 / NBRC 100331 / OCM 82 / Marburg References: UniProt: P11558, coenzyme-B sulfoethylthiotransferase #2: Protein | Mass: 47148.477 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Methanothermobacter marburgensis str. Marburg (archaea) Methanothermobacter marburgensis str. Marburg (archaea)Strain: ATCC BAA-927 / DSM 2133 / JCM 14651 / NBRC 100331 / OCM 82 / Marburg References: UniProt: P11560, coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 28423.768 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Methanothermobacter marburgensis str. Marburg (archaea) Methanothermobacter marburgensis str. Marburg (archaea)Strain: ATCC BAA-927 / DSM 2133 / JCM 14651 / NBRC 100331 / OCM 82 / Marburg References: UniProt: P11562, coenzyme-B sulfoethylthiotransferase |
|---|
-Non-polymers , 7 types, 749 molecules 
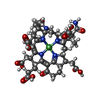











| #4: Chemical | ChemComp-MG / #5: Chemical | #6: Chemical | #7: Chemical | #8: Chemical | ChemComp-ACT / | #9: Chemical | ChemComp-EDO / | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 44.81 % Description: Pale yellow, Plate-like crystals ranging 20-60 microns in size |
|---|---|
| Crystal grow | Temperature: 300 K / Method: vapor diffusion, hanging drop / pH: 7.55 Details: 18% PEG 400, 150mM Mg Acetate, 100mM HEPES (pH 7.5) PH range: 7.5-8.0 / Temp details: Isotemp |
-Data collection
| Diffraction | Mean temperature: 300 K / Ambient temp details: Isotemp / Serial crystal experiment: Y |
|---|---|
| Diffraction source | Source:  FREE ELECTRON LASER / Site: FREE ELECTRON LASER / Site:  SLAC LCLS SLAC LCLS  / Beamline: MFX / Wavelength: 1.3007 Å / Beamline: MFX / Wavelength: 1.3007 Å |
| Detector | Type: RAYONIX MX340-HS / Detector: CCD / Date: Jul 11, 2020 / Frequency: 30 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.3007 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→21.25 Å / Num. obs: 189345 / % possible obs: 99.91 % / Redundancy: 41.43 % / Biso Wilson estimate: 26.7 Å2 / CC1/2: 0.979 / R split: 0.515 / Net I/σ(I): 3.356 |
| Reflection shell | Resolution: 1.9→1.93 Å / Redundancy: 9.89 % / Mean I/σ(I) obs: 0.696 / Num. unique obs: 9505 / CC1/2: 0.379 / R split: 0.831 / % possible all: 99.82 |
| Serial crystallography measurement | Collection time total: 0.68 hours / Focal spot size: 16 µm2 / Pulse duration: 30 fsec. / Pulse energy: 2200 µJ / Pulse photon energy: 9.5 keV / XFEL pulse repetition rate: 30 Hz |
| Serial crystallography sample delivery | Description: Drop On Tape / Method: injection |
| Serial crystallography sample delivery injection | Carrier solvent: mother liquor Description: Crystal slurry is injected onto a kapton tape from an acoustic droplet injector (ADE). The drop travels on the tape towards the X-ray interaction point where it is hit by the XFEL beam. ...Description: Crystal slurry is injected onto a kapton tape from an acoustic droplet injector (ADE). The drop travels on the tape towards the X-ray interaction point where it is hit by the XFEL beam. This is the Drop-On-Tape (DOT) sample delivery system used by our team previously. |
| Serial crystallography data reduction | Crystal hits: 24589 / Frames total: 73742 / Lattices indexed: 22706 / XFEL run numbers: 44-53 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3M1V Resolution: 1.9→21.25 Å / SU ML: 0.2017 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 18.6978 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.52 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→21.25 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj