+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7mdz | ||||||
|---|---|---|---|---|---|---|---|
| Title | 80S rabbit ribosome stalled with benzamide-CHX | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RIBOSOME / 80S mammalian ribosome / translation inhibitor | ||||||
| Function / homology |  Function and homology information Function and homology informationribosomal subunit / laminin receptor activity / ubiquitin ligase inhibitor activity / 90S preribosome / positive regulation of signal transduction by p53 class mediator / phagocytic cup / protein-RNA complex assembly / rough endoplasmic reticulum / laminin binding / ribosomal small subunit export from nucleus ...ribosomal subunit / laminin receptor activity / ubiquitin ligase inhibitor activity / 90S preribosome / positive regulation of signal transduction by p53 class mediator / phagocytic cup / protein-RNA complex assembly / rough endoplasmic reticulum / laminin binding / ribosomal small subunit export from nucleus / translation regulator activity / gastrulation / MDM2/MDM4 family protein binding / cytosolic ribosome / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of apoptotic signaling pathway / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / spindle / rRNA processing / positive regulation of canonical Wnt signaling pathway / rhythmic process / antimicrobial humoral immune response mediated by antimicrobial peptide / heparin binding / regulation of translation / large ribosomal subunit / ribosome binding / virus receptor activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / killing of cells of another organism / defense response to Gram-negative bacterium / perikaryon / cytoplasmic translation / cell differentiation / tRNA binding / mitochondrial inner membrane / rRNA binding / postsynaptic density / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / cell division / DNA repair / mRNA binding / apoptotic process / dendrite / synapse / centrosome / nucleolus / perinuclear region of cytoplasm / endoplasmic reticulum / Golgi apparatus / DNA binding / RNA binding / zinc ion binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||
 Authors Authors | Koga, Y. / Hoang, E.M. / Park, Y. / Keszei, A.F.A. / Murray, J. / Shao, S. / Liau, B.B. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J Am Chem Soc / Year: 2021 Journal: J Am Chem Soc / Year: 2021Title: Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation. Authors: Yumi Koga / Eileen M Hoang / Yongho Park / Alexander F A Keszei / Jason Murray / Sichen Shao / Brian B Liau /  Abstract: Employed for over half a century to study protein synthesis, cycloheximide (CHX, ) is a small molecule natural product that reversibly inhibits translation elongation. More recently, CHX has been ...Employed for over half a century to study protein synthesis, cycloheximide (CHX, ) is a small molecule natural product that reversibly inhibits translation elongation. More recently, CHX has been applied to ribosome profiling, a method for mapping ribosome positions on mRNA genome-wide. Despite CHX's extensive use, CHX treatment often results in incomplete translation inhibition due to its rapid reversibility, prompting the need for improved reagents. Here, we report the concise synthesis of C13-amide-functionalized CHX derivatives with increased potencies toward protein synthesis inhibition. Cryogenic electron microscopy (cryo-EM) revealed that C13-aminobenzoyl CHX () occupies the same site as CHX, competing with the 3' end of E-site tRNA. We demonstrate that is superior to CHX for ribosome profiling experiments, enabling more effective capture of ribosome conformations through sustained stabilization of polysomes. Our studies identify powerful chemical reagents to study protein synthesis and reveal the molecular basis of their enhanced potency. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7mdz.cif.gz 7mdz.cif.gz | 4.6 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7mdz.ent.gz pdb7mdz.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7mdz.json.gz 7mdz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/md/7mdz https://data.pdbj.org/pub/pdb/validation_reports/md/7mdz ftp://data.pdbj.org/pub/pdb/validation_reports/md/7mdz ftp://data.pdbj.org/pub/pdb/validation_reports/md/7mdz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  23785MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Ribosomal protein ... , 7 types, 7 molecules ANQRVjDD
| #1: Protein | Mass: 28088.863 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #13: Protein | Mass: 24207.285 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #16: Protein | Mass: 21699.688 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #17: Protein | Mass: 23535.281 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #21: Protein | Mass: 14892.505 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #35: Protein | Mass: 11111.032 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #46: Protein | Mass: 26715.344 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  References: UniProt: G1TNM3, DNA-(apurinic or apyrimidinic site) lyase |
+Protein , 45 types, 45 molecules BDOPTUXYcdefhlmoprBBCCFFGGHHIIJJKKLLMMNNOO...
-60S ribosomal protein ... , 18 types, 18 molecules CEFGHIJLMSWZabgikn
| #3: Protein | Mass: 47727.559 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #5: Protein | Mass: 33028.336 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #6: Protein | Mass: 29201.836 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #7: Protein | Mass: 36221.516 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #8: Protein | Mass: 21871.418 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #9: Protein | Mass: 24643.057 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #10: Protein | Mass: 20288.465 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #11: Protein | Mass: 24331.723 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #12: Protein | Mass: 23870.549 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #18: Protein | Mass: 20827.561 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #22: Protein | Mass: 17825.111 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #25: Protein | Mass: 15835.831 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #26: Protein | Mass: 16620.561 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #27: Protein | Mass: 26708.707 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #32: Protein | Mass: 13196.894 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #34: Protein | Mass: 12263.834 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #36: Protein | Mass: 8238.948 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #39: Protein/peptide | Mass: 3473.451 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-40S ribosomal protein ... , 5 types, 5 molecules AAEEVVYYaa
| #43: Protein | Mass: 32958.016 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #47: Protein | Mass: 29654.869 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #64: Protein | Mass: 9124.389 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #67: Protein | Mass: 15107.924 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #69: Protein | Mass: 13047.532 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-RNA chain , 6 types, 6 molecules 578296
| #76: RNA chain | Mass: 1167622.625 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #77: RNA chain | Mass: 38691.914 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #78: RNA chain | Mass: 50143.648 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #79: RNA chain | Mass: 24485.539 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #80: RNA chain | Mass: 602777.875 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #81: RNA chain | Mass: 3139.876 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host:  |
-Non-polymers , 3 types, 286 molecules 

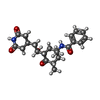


| #82: Chemical | ChemComp-MG / #83: Chemical | ChemComp-ZN / #84: Chemical | ChemComp-Z2V / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: 80S rabbit ribosome stalled with benzamide-CHX / Type: RIBOSOME / Entity ID: #1-#81 / Source: MULTIPLE SOURCES |
|---|---|
| Molecular weight | Value: 4 MDa / Experimental value: NO |
| Source (natural) | Organism:  |
| Buffer solution | pH: 7.4 |
| Specimen | Conc.: 0.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: Quantifoil |
| Vitrification | Instrument: FEI VITROBOT MARK II / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: 3 sec blot |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 47 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software | Name: RELION / Category: image acquisition |
|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Symmetry | Point symmetry: C1 (asymmetric) |
| 3D reconstruction | Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 34783 / Symmetry type: POINT |
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL |
| Atomic model building | PDB-ID: 6SGC Accession code: 6SGC / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller

















 PDBj
PDBj


































