[English] 日本語
 Yorodumi
Yorodumi- PDB-6tko: Phosphorylated turkey beta1 adrenoceptor with bound agonist formo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6tko | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Phosphorylated turkey beta1 adrenoceptor with bound agonist formoterol coupled to arrestin-2 in lipid nanodisc. | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | SIGNALING PROTEIN / GPCR / Arrestin / Complex / Nanodisc | |||||||||
| Function / homology |  Function and homology information Function and homology informationbeta1-adrenergic receptor activity / positive regulation of heart contraction / angiotensin receptor binding / regulation of circadian sleep/wake cycle, sleep / TGFBR3 regulates TGF-beta signaling / Activation of SMO / negative regulation of interleukin-8 production / arrestin family protein binding / G protein-coupled receptor internalization / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure ...beta1-adrenergic receptor activity / positive regulation of heart contraction / angiotensin receptor binding / regulation of circadian sleep/wake cycle, sleep / TGFBR3 regulates TGF-beta signaling / Activation of SMO / negative regulation of interleukin-8 production / arrestin family protein binding / G protein-coupled receptor internalization / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / Lysosome Vesicle Biogenesis / : / positive regulation of cardiac muscle hypertrophy / stress fiber assembly / Golgi Associated Vesicle Biogenesis / positive regulation of Rho protein signal transduction / pseudopodium / negative regulation of interleukin-6 production / positive regulation of receptor internalization / negative regulation of Notch signaling pathway / enzyme inhibitor activity / adenylate cyclase-activating adrenergic receptor signaling pathway / insulin-like growth factor receptor binding / clathrin-coated pit / negative regulation of protein ubiquitination / GTPase activator activity / Activated NOTCH1 Transmits Signal to the Nucleus / cytoplasmic vesicle membrane / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / G protein-coupled receptor binding / positive regulation of protein phosphorylation / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / endocytic vesicle membrane / Signaling by BRAF and RAF1 fusions / Cargo recognition for clathrin-mediated endocytosis / protein transport / Clathrin-mediated endocytosis / Thrombin signalling through proteinase activated receptors (PARs) / cytoplasmic vesicle / ubiquitin-dependent protein catabolic process / G alpha (s) signalling events / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / transcription coactivator activity / early endosome / positive regulation of ERK1 and ERK2 cascade / positive regulation of MAPK cascade / Ub-specific processing proteases / nuclear body / protein ubiquitination / Golgi membrane / lysosomal membrane / ubiquitin protein ligase binding / regulation of transcription by RNA polymerase II / chromatin / signal transduction / positive regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding / nucleus / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)Phage display vector pTDisp (others) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Lee, Y. / Tate, C.G. | |||||||||
| Funding support |  United Kingdom, 2items United Kingdom, 2items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Molecular basis of β-arrestin coupling to formoterol-bound β-adrenoceptor. Authors: Yang Lee / Tony Warne / Rony Nehmé / Shubhi Pandey / Hemlata Dwivedi-Agnihotri / Madhu Chaturvedi / Patricia C Edwards / Javier García-Nafría / Andrew G W Leslie / Arun K Shukla / Christopher G Tate /     Abstract: The β-adrenoceptor (βAR) is a G-protein-coupled receptor (GPCR) that couples to the heterotrimeric G protein G. G-protein-mediated signalling is terminated by phosphorylation of the C terminus of ...The β-adrenoceptor (βAR) is a G-protein-coupled receptor (GPCR) that couples to the heterotrimeric G protein G. G-protein-mediated signalling is terminated by phosphorylation of the C terminus of the receptor by GPCR kinases (GRKs) and by coupling of β-arrestin 1 (βarr1, also known as arrestin 2), which displaces G and induces signalling through the MAP kinase pathway. The ability of synthetic agonists to induce signalling preferentially through either G proteins or arrestins-known as biased agonism-is important in drug development, because the therapeutic effect may arise from only one signalling cascade, whereas the other pathway may mediate undesirable side effects. To understand the molecular basis for arrestin coupling, here we determined the cryo-electron microscopy structure of the βAR-βarr1 complex in lipid nanodiscs bound to the biased agonist formoterol, and the crystal structure of formoterol-bound βAR coupled to the G-protein-mimetic nanobody Nb80. βarr1 couples to βAR in a manner distinct to that of G coupling to βAR-the finger loop of βarr1 occupies a narrower cleft on the intracellular surface, and is closer to transmembrane helix H7 of the receptor when compared with the C-terminal α5 helix of G. The conformation of the finger loop in βarr1 is different from that adopted by the finger loop of visual arrestin when it couples to rhodopsin. βAR coupled to βarr1 shows considerable differences in structure compared with βAR coupled to Nb80, including an inward movement of extracellular loop 3 and the cytoplasmic ends of H5 and H6. We observe weakened interactions between formoterol and two serine residues in H5 at the orthosteric binding site of βAR, and find that formoterol has a lower affinity for the βAR-βarr1 complex than for the βAR-G complex. The structural differences between these complexes of βAR provide a foundation for the design of small molecules that could bias signalling in the β-adrenoceptors. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Molecular determinants of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor. Authors: Lee, Y. / Warne, T. / Nehme, R. / Pandey, S. / Dwivedi-Agnihotri, H. / Edwards, P.C. / Garcia-Nafria, J. / Leslie, A.G.W. / Shukla, A.K. / Tate, C.G. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6tko.cif.gz 6tko.cif.gz | 205.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6tko.ent.gz pdb6tko.ent.gz | 152.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6tko.json.gz 6tko.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6tko_validation.pdf.gz 6tko_validation.pdf.gz | 1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6tko_full_validation.pdf.gz 6tko_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  6tko_validation.xml.gz 6tko_validation.xml.gz | 40.2 KB | Display | |
| Data in CIF |  6tko_validation.cif.gz 6tko_validation.cif.gz | 62.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tk/6tko https://data.pdbj.org/pub/pdb/validation_reports/tk/6tko ftp://data.pdbj.org/pub/pdb/validation_reports/tk/6tko ftp://data.pdbj.org/pub/pdb/validation_reports/tk/6tko | HTTPS FTP |
-Related structure data
| Related structure data |  10515MC  6iblC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10342 (Title: Cryo-EM structure of the formoterol-bound turkey beta1 adrenoceptor coupled to beta-arrestin-1 with bound Fab30 in lipid nanodisc EMPIAR-10342 (Title: Cryo-EM structure of the formoterol-bound turkey beta1 adrenoceptor coupled to beta-arrestin-1 with bound Fab30 in lipid nanodiscData size: 4.0 TB Data #1: Unaligned multi-frame micrographs [micrographs - multiframe] Data #2: Unaligned multi-frame micrographs [micrographs - multiframe] Data #3: Unaligned multi-frame micrographs [micrographs - multiframe] Data #4: Nanodisc subtracted particles [picked particles - multiframe - processed] Data #5: Nanodisc-reinstated particles [picked particles - multiframe - unprocessed]) |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 37275.395 Da / Num. of mol.: 1 Mutation: S32G M44C M90V V103C C116L E130W D322K F327A F338M C358A Source method: isolated from a genetically manipulated source Details: Relative to wild-type sequence: construct is truncated at the N-terminus; and in ICL3; its C-terminus is truncated at C358A and fused with a linker to a series of phosphorylated residues ...Details: Relative to wild-type sequence: construct is truncated at the N-terminus; and in ICL3; its C-terminus is truncated at C358A and fused with a linker to a series of phosphorylated residues (ARGRPLPETGGGDE[pS]A[pT][pT]A[pS][pS][pS]LAKDTSS). Source: (gene. exp.)   Trichoplusia ni (cabbage looper) / References: UniProt: P07700 Trichoplusia ni (cabbage looper) / References: UniProt: P07700 |
|---|---|
| #2: Protein | Mass: 47021.332 Da / Num. of mol.: 1 / Mutation: M1G L68C R169E Source method: isolated from a genetically manipulated source Details: Additional N-terminal Gly residue, in place of starting Met, remaining from proteolytic cleavage. Source: (gene. exp.)  Homo sapiens (human) / Gene: ARRB1, ARR1 / Production host: Homo sapiens (human) / Gene: ARRB1, ARR1 / Production host:  |
| #3: Antibody | Mass: 25512.354 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) Phage display vector pTDisp (others) / Production host:  |
| #4: Antibody | Mass: 23435.064 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) Phage display vector pTDisp (others) / Production host:  |
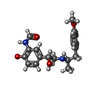
| #5: Chemical | ChemComp-H98 / ~{ |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.133 MDa / Experimental value: NO | ||||||||||||||||||||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: This sample was monodisperse. | ||||||||||||||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K Details: Blotted for 2-3 seconds before plunging. Liquid ethane maintained at 92.15 K. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM imaging | Accelerating voltage: 300 kV / Alignment procedure: COMA FREE / C2 aperture diameter: 100 µm / Cryogen: NITROGEN / Electron source:
| |||||||||||||||
| Image recording |
| |||||||||||||||
| EM imaging optics |
|
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 10161197 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 403991 Details: Half maps were locally filtered between refinement iterations using SIDESPLITTER, an adaptation of the LAFTER algorithm that maintains gold-standard separation between the two half maps. Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 80.6 / Protocol: OTHER / Space: REAL / Target criteria: Correlation coefficient Details: Initial placement was done manually in Coot, followed by rigid-body refinement in PHENIX and jelly-body refinement in REFMAC5. Manual remodeling was performed in Coot and iterated with real ...Details: Initial placement was done manually in Coot, followed by rigid-body refinement in PHENIX and jelly-body refinement in REFMAC5. Manual remodeling was performed in Coot and iterated with real space refinement in PHENIX. Chemical restraints for formoterol were calculated in eLBOW using AM1 optimisation. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 80.62 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera






 PDBj
PDBj