+Search query
-Structure paper
| Title | Molecular basis of β-arrestin coupling to formoterol-bound β-adrenoceptor. |
|---|---|
| Journal, issue, pages | Nature, Vol. 583, Issue 7818, Page 862-866, Year 2020 |
| Publish date | Jun 17, 2020 |
 Authors Authors | Yang Lee / Tony Warne / Rony Nehmé / Shubhi Pandey / Hemlata Dwivedi-Agnihotri / Madhu Chaturvedi / Patricia C Edwards / Javier García-Nafría / Andrew G W Leslie / Arun K Shukla / Christopher G Tate /     |
| PubMed Abstract | The β-adrenoceptor (βAR) is a G-protein-coupled receptor (GPCR) that couples to the heterotrimeric G protein G. G-protein-mediated signalling is terminated by phosphorylation of the C terminus of ...The β-adrenoceptor (βAR) is a G-protein-coupled receptor (GPCR) that couples to the heterotrimeric G protein G. G-protein-mediated signalling is terminated by phosphorylation of the C terminus of the receptor by GPCR kinases (GRKs) and by coupling of β-arrestin 1 (βarr1, also known as arrestin 2), which displaces G and induces signalling through the MAP kinase pathway. The ability of synthetic agonists to induce signalling preferentially through either G proteins or arrestins-known as biased agonism-is important in drug development, because the therapeutic effect may arise from only one signalling cascade, whereas the other pathway may mediate undesirable side effects. To understand the molecular basis for arrestin coupling, here we determined the cryo-electron microscopy structure of the βAR-βarr1 complex in lipid nanodiscs bound to the biased agonist formoterol, and the crystal structure of formoterol-bound βAR coupled to the G-protein-mimetic nanobody Nb80. βarr1 couples to βAR in a manner distinct to that of G coupling to βAR-the finger loop of βarr1 occupies a narrower cleft on the intracellular surface, and is closer to transmembrane helix H7 of the receptor when compared with the C-terminal α5 helix of G. The conformation of the finger loop in βarr1 is different from that adopted by the finger loop of visual arrestin when it couples to rhodopsin. βAR coupled to βarr1 shows considerable differences in structure compared with βAR coupled to Nb80, including an inward movement of extracellular loop 3 and the cytoplasmic ends of H5 and H6. We observe weakened interactions between formoterol and two serine residues in H5 at the orthosteric binding site of βAR, and find that formoterol has a lower affinity for the βAR-βarr1 complex than for the βAR-G complex. The structural differences between these complexes of βAR provide a foundation for the design of small molecules that could bias signalling in the β-adrenoceptors. |
 External links External links |  Nature / Nature /  PubMed:32555462 / PubMed:32555462 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.7 - 3.3 Å |
| Structure data | EMDB-10515, PDB-6tko:  PDB-6ibl: |
| Chemicals | 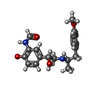 ChemComp-H98:  ChemComp-NA: 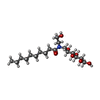 ChemComp-2CV:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / Beta1 Adrenoceptor / Activated / Agonist / Nanobody / SIGNALING PROTEIN / GPCR / Arrestin / Complex / Nanodisc |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






 homo sapiens (human)
homo sapiens (human)

