[English] 日本語
 Yorodumi
Yorodumi- PDB-6qox: Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6qox | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 27 (Methyl 2-(hydroxymethyl)-6H-thieno[2,3-b]pyrrole-5-carboxylate) | ||||||
 Components Components | tRNA (guanine-N(1)-)-methyltransferase | ||||||
 Keywords Keywords | TRANSFERASE / TrmD / tRNA methyltransferase / SPOUT methyltransferase | ||||||
| Function / homology |  Function and homology information Function and homology informationtRNA (guanine37-N1)-methyltransferase / tRNA (guanine(37)-N1)-methyltransferase activity / tRNA modification / methylation / cytoplasm Similarity search - Function | ||||||
| Biological species |  Mycobacterium abscessus (bacteria) Mycobacterium abscessus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.74 Å molecular replacement / Resolution: 1.74 Å | ||||||
 Authors Authors | Thomas, S.E. / Whitehouse, A.J. / Coyne, A.G. / Abell, C. / Mendes, V. / Blundell, T.L. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2020 Journal: Nucleic Acids Res. / Year: 2020Title: Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification. Authors: Thomas, S.E. / Whitehouse, A.J. / Brown, K. / Burbaud, S. / Belardinelli, J.M. / Sangen, J. / Lahiri, R. / Libardo, M.D.J. / Gupta, P. / Malhotra, S. / Boshoff, H.I.M. / Jackson, M. / Abell, ...Authors: Thomas, S.E. / Whitehouse, A.J. / Brown, K. / Burbaud, S. / Belardinelli, J.M. / Sangen, J. / Lahiri, R. / Libardo, M.D.J. / Gupta, P. / Malhotra, S. / Boshoff, H.I.M. / Jackson, M. / Abell, C. / Coyne, A.G. / Blundell, T.L. / Floto, R.A. / Mendes, V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6qox.cif.gz 6qox.cif.gz | 184.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6qox.ent.gz pdb6qox.ent.gz | 145.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6qox.json.gz 6qox.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6qox_validation.pdf.gz 6qox_validation.pdf.gz | 443.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6qox_full_validation.pdf.gz 6qox_full_validation.pdf.gz | 444.3 KB | Display | |
| Data in XML |  6qox_validation.xml.gz 6qox_validation.xml.gz | 19.7 KB | Display | |
| Data in CIF |  6qox_validation.cif.gz 6qox_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qo/6qox https://data.pdbj.org/pub/pdb/validation_reports/qo/6qox ftp://data.pdbj.org/pub/pdb/validation_reports/qo/6qox ftp://data.pdbj.org/pub/pdb/validation_reports/qo/6qox | HTTPS FTP |
-Related structure data
| Related structure data |  6nvrC  6nw6C  6nw7C  6qo2C  6qo3C  6qo4C  6qo6C  6qoaC  6qocC  6qodC  6qoeC  6qofC  6qogC  6qohC  6qoiC  6qojC  6qokC  6qolC  6qomC  6qonC  6qooC  6qopC  6qoqC  6qorC  6qosC  6qotC  6qouC  6qovC  6qowC  6qqsC  6qqxC  6qqyC  6qr5C  6qr6C  6qr8C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
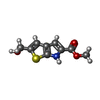
| #1: Protein | Mass: 26434.670 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mycobacterium abscessus (bacteria) / Gene: trmD, MAB_3226c / Production host: Mycobacterium abscessus (bacteria) / Gene: trmD, MAB_3226c / Production host:  References: UniProt: B1MDI3, tRNA (guanine37-N1)-methyltransferase #2: Chemical | ChemComp-JA5 / | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 50.02 % |
|---|---|
| Crystal grow | Temperature: 292 K / Method: vapor diffusion, sitting drop Details: 0.08M Sodium cacodylate pH 6.5 -7.0, 1-2 M Ammonium sulphate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.9282 Å / Beamline: I04-1 / Wavelength: 0.9282 Å | ||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Dec 14, 2015 | ||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9282 Å / Relative weight: 1 | ||||||||||||||||||||||||
| Reflection | Resolution: 1.74→88.48 Å / Num. obs: 54370 / % possible obs: 100 % / Redundancy: 7.8 % / Biso Wilson estimate: 29.03 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.044 / Rpim(I) all: 0.017 / Rrim(I) all: 0.047 / Net I/σ(I): 23.5 | ||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.74→58.705 Å / SU ML: 0.2 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 21.18 MOLECULAR REPLACEMENT / Resolution: 1.74→58.705 Å / SU ML: 0.2 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 21.18
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 96.55 Å2 / Biso mean: 36.9592 Å2 / Biso min: 18.27 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.74→58.705 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 20 / % reflection obs: 100 %
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj