| Entry | Database: PDB / ID: 6q5m
|
|---|
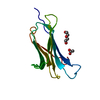
| Title | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24Dab |
|---|
 Components Components | CC-Hex*-L24Dab  Keywords Keywords | DE NOVO PROTEIN / coiled coil / tetramer / synthetic / antiparallel / CC-Hex |
|---|
| Biological species | synthetic construct (others) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å |
|---|
 Authors Authors | Wood, C.W. / Beesley, J.L. / Rhys, G.G. / Brady, R.L. / Woolfson, D.N. |
|---|
| Funding support |  United Kingdom, United Kingdom,  Belgium, 5items Belgium, 5items | Organization | Grant number | Country |
|---|
| Biotechnology and Biological Sciences Research Council | BB/J014400/1 |  United Kingdom United Kingdom | | Engineering and Physical Sciences Research Council | EP/G036764/1 |  United Kingdom United Kingdom | | European Research Council | 340764 |  Belgium Belgium | | Biotechnology and Biological Sciences Research Council | BB/R00661X/1 |  United Kingdom United Kingdom | | Engineering and Physical Sciences Research Council | EP/K03927X/1 |  United Kingdom United Kingdom |
|
|---|
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2019 Journal: J.Am.Chem.Soc. / Year: 2019
Title: Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
Authors: Rhys, G.G. / Wood, C.W. / Beesley, J.L. / Zaccai, N.R. / Burton, A.J. / Brady, R.L. / Thomson, A.R. / Woolfson, D.N. |
|---|
| History | | Deposition | Dec 9, 2018 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | May 22, 2019 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jun 19, 2019 | Group: Data collection / Database references / Category: citation / citation_author / pdbx_database_proc
Item: _citation.journal_volume / _citation.page_first ..._citation.journal_volume / _citation.page_first / _citation.page_last / _citation.title / _citation_author.identifier_ORCID / _citation_author.name |
|---|
| Revision 1.2 | Oct 16, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry |
|---|
|
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å
MOLECULAR REPLACEMENT / Resolution: 1.5 Å  Authors
Authors United Kingdom,
United Kingdom,  Belgium, 5items
Belgium, 5items  Citation
Citation Journal: J.Am.Chem.Soc. / Year: 2019
Journal: J.Am.Chem.Soc. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6q5m.cif.gz
6q5m.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6q5m.ent.gz
pdb6q5m.ent.gz PDB format
PDB format 6q5m.json.gz
6q5m.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/q5/6q5m
https://data.pdbj.org/pub/pdb/validation_reports/q5/6q5m ftp://data.pdbj.org/pub/pdb/validation_reports/q5/6q5m
ftp://data.pdbj.org/pub/pdb/validation_reports/q5/6q5m









 Links
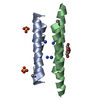
Links Assembly
Assembly

 Components
Components X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I03 / Wavelength: 0.9783 Å
/ Beamline: I03 / Wavelength: 0.9783 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 1.5→35.72 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.95 / SU B: 1.944 / SU ML: 0.067 / Cross valid method: THROUGHOUT / ESU R: 0.078 / ESU R Free: 0.081 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 1.5→35.72 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.95 / SU B: 1.944 / SU ML: 0.067 / Cross valid method: THROUGHOUT / ESU R: 0.078 / ESU R Free: 0.081 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller











 PDBj
PDBj


