[English] 日本語
 Yorodumi
Yorodumi- PDB-1mol: TWO CRYSTAL STRUCTURES OF A POTENTLY SWEET PROTEIN: NATURAL MONEL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mol | ||||||
|---|---|---|---|---|---|---|---|
| Title | TWO CRYSTAL STRUCTURES OF A POTENTLY SWEET PROTEIN: NATURAL MONELLIN AT 2.75 ANGSTROMS RESOLUTION AND SINGLE-CHAIN MONELLIN AT 1.7 ANGSTROMS RESOLUTION | ||||||
 Components Components | MONELLIN | ||||||
 Keywords Keywords | SWEET-TASTING PROTEIN | ||||||
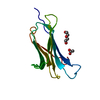
| Function / homology |  Function and homology information Function and homology informationMonellin, B chain / : / Monellin / Monellin / Nuclear Transport Factor 2; Chain: A, - #10 / Cystatin superfamily / Nuclear Transport Factor 2; Chain: A, / Roll / Alpha Beta Similarity search - Domain/homology | ||||||
| Biological species |  Dioscoreophyllum cumminsii (serendipity berry) Dioscoreophyllum cumminsii (serendipity berry) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.7 Å X-RAY DIFFRACTION / Resolution: 1.7 Å | ||||||
 Authors Authors | Somoza, J.R. / Kim, S.-H. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: Two crystal structures of a potently sweet protein. Natural monellin at 2.75 A resolution and single-chain monellin at 1.7 A resolution. Authors: Somoza, J.R. / Jiang, F. / Tong, L. / Kang, C.H. / Cho, J.M. / Kim, S.H. #1:  Journal: J.Biomol.NMR / Year: 1992 Journal: J.Biomol.NMR / Year: 1992Title: 1H Resonance Assignments, Secondary Structure and General Topology of Single-Chain Monellin in Solution as Determined by 1H-2D-NMR Authors: Tomic, M.T. / Somoza, J.R. / Wemmer, D.E. / Park, Y.W. / Cho, J.M. / Kim, S.-H. #2:  Journal: Protein Eng. / Year: 1989 Journal: Protein Eng. / Year: 1989Title: Redesigning a Sweet Protein: Increased Stability and Renaturability Authors: Kim, S.-H. / Kang, C.-H. / Kim, R. / Cho, J.M. / Lee, Y.-B. / Lee, T.-K. | ||||||
| History |
| ||||||
| Remark 650 | HELIX THERE IS A SINGLE RIGHT-HANDED ALPHA-HELIX IN EACH OF THE MONOMERS PRESENT IN THE ASYMMETRIC UNIT. | ||||||
| Remark 700 | SHEET EACH OF THE MONOMERS IN THE ASYMMETRIC UNIT CONTAINS A SINGLE, FIVE-STRAND ANTIPARALLEL BETA-SHEET. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mol.cif.gz 1mol.cif.gz | 52 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mol.ent.gz pdb1mol.ent.gz | 38.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mol.json.gz 1mol.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mo/1mol https://data.pdbj.org/pub/pdb/validation_reports/mo/1mol ftp://data.pdbj.org/pub/pdb/validation_reports/mo/1mol ftp://data.pdbj.org/pub/pdb/validation_reports/mo/1mol | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO A 40 / 2: CIS PROLINE - PRO A 90 / 3: CIS PROLINE - PRO B 40 / 4: CIS PROLINE - PRO B 90 |
- Components
Components
| #1: Protein | Mass: 11083.628 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Dioscoreophyllum cumminsii (serendipity berry) Dioscoreophyllum cumminsii (serendipity berry)References: UniProt: P02882 #2: Water | ChemComp-HOH / | Sequence details | SEQUENCE ADVISORY NOTICE DIFFERENCE BETWEEN SWISS-PROT AND PDB SEQUENCE. SWISS-PROT ENTRY NAME: ...SEQUENCE ADVISORY NOTICE DIFFERENCE | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 39.61 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 7.2 / Method: vapor diffusion | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.6 Å / Num. obs: 19162 / % possible obs: 82.9 % / Observed criterion σ(I): 0 / Num. measured all: 59663 / Rmerge(I) obs: 0.0638 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.7→6 Å / σ(F): 2 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Num. reflection all: 17089 / Rfactor all: 0.188 / Rfactor obs: 0.174 / Rfactor Rwork: 0.174 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 23.6 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: x_angle_d / Dev ideal: 2.86 |
 Movie
Movie Controller
Controller











 PDBj
PDBj

