[English] 日本語
 Yorodumi
Yorodumi- PDB-6ibq: Structure of a nonameric RNA duplex at room temperature in ChipX ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ibq | ||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
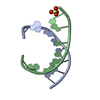
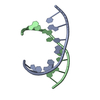
| Title | Structure of a nonameric RNA duplex at room temperature in ChipX microfluidic device | ||||||||||||||||||||||||||||||||||
 Components Components | DNA/RNA (5'-R(* Keywords KeywordsRNA / RNA duplex / GoU pair / chipx | Function / homology | DNA/RNA hybrid |  Function and homology information Function and homology informationBiological species |  Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.55 Å MOLECULAR REPLACEMENT / Resolution: 1.55 Å  Authors Authorsde Wijn, R. / Olieric, V. / Lorber, B. / Sauter, C. | Funding support | |  France, 3items France, 3items
 Citation Citation Journal: Iucrj / Year: 2019 Journal: Iucrj / Year: 2019Title: A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography. Authors: de Wijn, R. / Hennig, O. / Roche, J. / Engilberge, S. / Rollet, K. / Fernandez-Millan, P. / Brillet, K. / Betat, H. / Morl, M. / Roussel, A. / Girard, E. / Mueller-Dieckmann, C. / Fox, G.C. ...Authors: de Wijn, R. / Hennig, O. / Roche, J. / Engilberge, S. / Rollet, K. / Fernandez-Millan, P. / Brillet, K. / Betat, H. / Morl, M. / Roussel, A. / Girard, E. / Mueller-Dieckmann, C. / Fox, G.C. / Olieric, V. / Gavira, J.A. / Lorber, B. / Sauter, C. #1:  Journal: RNA / Year: 1999 Journal: RNA / Year: 1999Title: A sulfate pocket formed by three GoU pairs in the 0.97 A resolution X-ray structure of a nonameric RNA. Authors: Masquida, B. / Sauter, C. / Westhof, E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ibq.cif.gz 6ibq.cif.gz | 21.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ibq.ent.gz pdb6ibq.ent.gz | 12.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ibq.json.gz 6ibq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6ibq_validation.pdf.gz 6ibq_validation.pdf.gz | 374.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6ibq_full_validation.pdf.gz 6ibq_full_validation.pdf.gz | 374.6 KB | Display | |
| Data in XML |  6ibq_validation.xml.gz 6ibq_validation.xml.gz | 3.1 KB | Display | |
| Data in CIF |  6ibq_validation.cif.gz 6ibq_validation.cif.gz | 3.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ib/6ibq https://data.pdbj.org/pub/pdb/validation_reports/ib/6ibq ftp://data.pdbj.org/pub/pdb/validation_reports/ib/6ibq ftp://data.pdbj.org/pub/pdb/validation_reports/ib/6ibq | HTTPS FTP |
-Related structure data
| Related structure data |  6gzpC  6hw1C  6ibpC  6q3tC  6q52C  485dS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 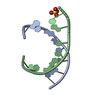
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA/RNA hybrid | Mass: 2831.743 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  #2: Chemical | ChemComp-SO4 / | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.83 Å3/Da / Density % sol: 54.7 % |
|---|---|
| Crystal grow | Temperature: 300 K / Method: counter-diffusion / pH: 6 Details: RNA solution: 10 mg/mL in 10 mM Na-cacodylate pH 6.0, 5 mM MgCl2. Reservoir solution: 2.6 M ammonium sulfate, 50 mM Na-cacodylate pH 6.0, 5 mM MgSO4, 1 mM spermine. Crystallization and ...Details: RNA solution: 10 mg/mL in 10 mM Na-cacodylate pH 6.0, 5 mM MgCl2. Reservoir solution: 2.6 M ammonium sulfate, 50 mM Na-cacodylate pH 6.0, 5 mM MgSO4, 1 mM spermine. Crystallization and crystallographic analysis were performed using the ChipX microfluidic device. |
-Data collection
| Diffraction | Mean temperature: 293 K / Ambient temp details: in situ / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 Å / Beamline: X06DA / Wavelength: 1 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Aug 31, 2011 |
| Radiation | Monochromator: Bartel monochromator with dual channel cut crystals (DCCM) Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.55→24 Å / Num. obs: 5485 / % possible obs: 91.5 % / Observed criterion σ(I): -3 / Redundancy: 3.9 % / Biso Wilson estimate: 23.6 Å2 / CC1/2: 0.988 / Rmerge(I) obs: 0.157 / Rrim(I) all: 0.179 / Net I/σ(I): 6.1 |
| Reflection shell | Resolution: 1.55→1.59 Å / Redundancy: 2 % / Rmerge(I) obs: 0.367 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 304 / CC1/2: 0.755 / Rrim(I) all: 0.455 / % possible all: 69.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 485D Resolution: 1.55→23.037 Å / Cross valid method: THROUGHOUT / σ(F): 2.55 / Phase error: 25.79 Details: The structure was refined using data collected on four crystals at room temperature. The first nucleotide of each chain was not visible in the electron density and is not included in the model.
| ||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.55→23.037 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj


