+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6hb4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | TFAM in Complex with Site-Y | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN / HMG-box / Transcription activator / Mitochondrial DNA / mtDNA Control region / DNA Compaction | ||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial transcription initiation / mitochondrial respiratory chain complex assembly / mitochondrial transcription factor activity / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / mitochondrial transcription / DNA binding, bending / mitochondrial nucleoid / heat shock protein binding / response to nutrient ...Mitochondrial transcription initiation / mitochondrial respiratory chain complex assembly / mitochondrial transcription factor activity / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / mitochondrial transcription / DNA binding, bending / mitochondrial nucleoid / heat shock protein binding / response to nutrient / Mitochondrial protein degradation / Transcriptional activation of mitochondrial biogenesis / transcription coactivator binding / sequence-specific DNA binding / response to hypoxia / transcription cis-regulatory region binding / mitochondrial matrix / chromatin binding / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / protein-containing complex / mitochondrion / RNA binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.05 Å MOLECULAR REPLACEMENT / Resolution: 3.05 Å | ||||||
 Authors Authors | Cuppari, A. / Fernandez-Millan, P. / Rubio-Cosials, A. / Tarres-Sole, A. / Lyonnais, S. / Sola, M. | ||||||
| Funding support |  Spain, 1items Spain, 1items
| ||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2019 Journal: Nucleic Acids Res. / Year: 2019Title: DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region. Authors: Cuppari, A. / Fernandez-Millan, P. / Battistini, F. / Tarres-Sole, A. / Lyonnais, S. / Iruela, G. / Ruiz-Lopez, E. / Enciso, Y. / Rubio-Cosials, A. / Prohens, R. / Pons, M. / Alfonso, C. / ...Authors: Cuppari, A. / Fernandez-Millan, P. / Battistini, F. / Tarres-Sole, A. / Lyonnais, S. / Iruela, G. / Ruiz-Lopez, E. / Enciso, Y. / Rubio-Cosials, A. / Prohens, R. / Pons, M. / Alfonso, C. / Toth, K. / Rivas, G. / Orozco, M. / Sola, M. #1:  Journal: Nat. Struct. Mol. Biol. / Year: 2011 Journal: Nat. Struct. Mol. Biol. / Year: 2011Title: Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Authors: Rubio-Cosials, A. / Sidow, J.F. / Jimenez-Menendez, N. / Fernandez-Millan, P. / Montoya, J. / Jacobs, H.T. / Coll, M. / Bernado, P. / Sola, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6hb4.cif.gz 6hb4.cif.gz | 529.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6hb4.ent.gz pdb6hb4.ent.gz | 435.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6hb4.json.gz 6hb4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hb/6hb4 https://data.pdbj.org/pub/pdb/validation_reports/hb/6hb4 ftp://data.pdbj.org/pub/pdb/validation_reports/hb/6hb4 ftp://data.pdbj.org/pub/pdb/validation_reports/hb/6hb4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 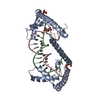 6hc3C  3tq6S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 25651.559 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TFAM, TCF6, TCF6L2 / Production host: Homo sapiens (human) / Gene: TFAM, TCF6, TCF6L2 / Production host:  #2: DNA chain | Mass: 6741.376 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: mitochondrial DNA / Source: (synth.)  Homo sapiens (human) Homo sapiens (human)#3: DNA chain | Mass: 6759.404 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: mitochondrial DNA / Source: (synth.)  Homo sapiens (human) Homo sapiens (human)#4: Chemical | #5: Chemical | ChemComp-1PE / |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.93 Å3/Da / Density % sol: 58.03 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion Details: 23-28% PEG 3350, 0.08-0.2M ammonium acetate, and 0.1 M bis-Tris pH 6.5 or Hepes pH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-2 / Wavelength: 0.93928 Å / Beamline: ID23-2 / Wavelength: 0.93928 Å |
| Detector | Type: DECTRIS PILATUS3 X 2M / Detector: PIXEL / Date: Sep 10, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.93928 Å / Relative weight: 1 |
| Reflection | Resolution: 3.05→43.7 Å / Num. obs: 34352 / % possible obs: 99.96 % / Redundancy: 13.6 % / Biso Wilson estimate: 81.52 Å2 / Rsym value: 0.103 / Net I/σ(I): 19.4 |
| Reflection shell | Resolution: 3.05→3.14 Å / Num. unique obs: 2796 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3TQ6 Resolution: 3.05→43.05 Å / Cor.coef. Fo:Fc: 0.9379 / Cor.coef. Fo:Fc free: 0.9037 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.384
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 88.24 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.57 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.05→43.05 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.05→3.14 Å / Total num. of bins used: 17
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj










































