+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 6g0q | ||||||
|---|---|---|---|---|---|---|---|
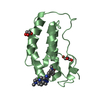
| タイトル | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード | TRANSCRIPTION / Bromodomain / complex | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of transcription regulatory region DNA binding / regulation of primitive erythrocyte differentiation / basophil differentiation / eosinophil fate commitment / regulation of definitive erythrocyte differentiation / eosinophil differentiation / regulation of glycoprotein biosynthetic process / primitive erythrocyte differentiation / myeloid cell apoptotic process / megakaryocyte differentiation ...negative regulation of transcription regulatory region DNA binding / regulation of primitive erythrocyte differentiation / basophil differentiation / eosinophil fate commitment / regulation of definitive erythrocyte differentiation / eosinophil differentiation / regulation of glycoprotein biosynthetic process / primitive erythrocyte differentiation / myeloid cell apoptotic process / megakaryocyte differentiation / osteoblast proliferation / Sertoli cell development / dendritic cell differentiation / negative regulation of bone mineralization / cellular response to follicle-stimulating hormone stimulus / positive regulation of mast cell degranulation / negative regulation of myeloid cell apoptotic process / C2H2 zinc finger domain binding / platelet formation / positive regulation of peptidyl-tyrosine phosphorylation / positive regulation of osteoblast proliferation / erythrocyte development / bone mineralization / RNA polymerase II C-terminal domain binding / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / host-mediated suppression of viral transcription / animal organ regeneration / cell fate commitment / positive regulation of G2/M transition of mitotic cell cycle / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of T-helper 17 cell lineage commitment / : / transcription repressor complex / RNA polymerase II CTD heptapeptide repeat kinase activity / homeostasis of number of cells within a tissue / positive regulation of erythrocyte differentiation / cellular response to cAMP / erythrocyte differentiation / condensed nuclear chromosome / transcription coregulator binding / transcription coregulator activity / RNA polymerase II transcription regulatory region sequence-specific DNA binding / positive regulation of transcription elongation by RNA polymerase II / protein-DNA complex / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / chromatin DNA binding / platelet aggregation / transcription coactivator binding / male gonad development / sequence-specific double-stranded DNA binding / p53 binding / cell-cell signaling / chromosome / RUNX1 regulates transcription of genes involved in differentiation of HSCs / cellular response to lipopolysaccharide / Factors involved in megakaryocyte development and platelet production / positive regulation of cytosolic calcium ion concentration / regulation of inflammatory response / DNA-binding transcription activator activity, RNA polymerase II-specific / histone binding / transcription regulator complex / sequence-specific DNA binding / in utero embryonic development / Potential therapeutics for SARS / RNA polymerase II-specific DNA-binding transcription factor binding / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription coactivator activity / positive regulation of canonical NF-kappaB signal transduction / transcription cis-regulatory region binding / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / DNA-binding transcription factor activity / negative regulation of cell population proliferation / protein serine/threonine kinase activity / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / chromatin / positive regulation of DNA-templated transcription / enzyme binding / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / nucleus 類似検索 - 分子機能 | ||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.4 Å 分子置換 / 解像度: 1.4 Å | ||||||
 データ登録者 データ登録者 | Filippakopoulos, P. / Picaud, S. / Newman, J. / Sorrell, F. / von Delft, F. / Arrowsmith, C.H. / Edwards, A.M. / Bountra, C. | ||||||
| 資金援助 |  英国, 1件 英国, 1件
| ||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2019 ジャーナル: Mol Cell / 年: 2019タイトル: Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains. 著者: Jean-Philippe Lambert / Sarah Picaud / Takao Fujisawa / Huayun Hou / Pavel Savitsky / Liis Uusküla-Reimand / Gagan D Gupta / Hala Abdouni / Zhen-Yuan Lin / Monika Tucholska / James D R ...著者: Jean-Philippe Lambert / Sarah Picaud / Takao Fujisawa / Huayun Hou / Pavel Savitsky / Liis Uusküla-Reimand / Gagan D Gupta / Hala Abdouni / Zhen-Yuan Lin / Monika Tucholska / James D R Knight / Beatriz Gonzalez-Badillo / Nicole St-Denis / Joseph A Newman / Manuel Stucki / Laurence Pelletier / Nuno Bandeira / Michael D Wilson / Panagis Filippakopoulos / Anne-Claude Gingras /      要旨: Targeting bromodomains (BRDs) of the bromo-and-extra-terminal (BET) family offers opportunities for therapeutic intervention in cancer and other diseases. Here, we profile the interactomes of BRD2, ...Targeting bromodomains (BRDs) of the bromo-and-extra-terminal (BET) family offers opportunities for therapeutic intervention in cancer and other diseases. Here, we profile the interactomes of BRD2, BRD3, BRD4, and BRDT following treatment with the pan-BET BRD inhibitor JQ1, revealing broad rewiring of the interaction landscape, with three distinct classes of behavior for the 603 unique interactors identified. A group of proteins associate in a JQ1-sensitive manner with BET BRDs through canonical and new binding modes, while two classes of extra-terminal (ET)-domain binding motifs mediate acetylation-independent interactions. Last, we identify an unexpected increase in several interactions following JQ1 treatment that define negative functions for BRD3 in the regulation of rRNA synthesis and potentially RNAPII-dependent gene expression that result in decreased cell proliferation. Together, our data highlight the contributions of BET protein modules to their interactomes allowing for a better understanding of pharmacological rewiring in response to JQ1. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  6g0q.cif.gz 6g0q.cif.gz | 81.8 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb6g0q.ent.gz pdb6g0q.ent.gz | 58.8 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  6g0q.json.gz 6g0q.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  6g0q_validation.pdf.gz 6g0q_validation.pdf.gz | 433.2 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  6g0q_full_validation.pdf.gz 6g0q_full_validation.pdf.gz | 433.3 KB | 表示 | |
| XML形式データ |  6g0q_validation.xml.gz 6g0q_validation.xml.gz | 9.9 KB | 表示 | |
| CIF形式データ |  6g0q_validation.cif.gz 6g0q_validation.cif.gz | 14.4 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/g0/6g0q https://data.pdbj.org/pub/pdb/validation_reports/g0/6g0q ftp://data.pdbj.org/pub/pdb/validation_reports/g0/6g0q ftp://data.pdbj.org/pub/pdb/validation_reports/g0/6g0q | HTTPS FTP |
-関連構造データ
| 関連構造データ |  5nncC  5nndC  5nneC  5nnfC  5nngC  6g0oC  6g0pC  6g0rC  6g0sC  2grcS  2oo1S  2ossS  2ouoS  3d7cS  3daiS  3dwyS S: 精密化の開始モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 単位格子 |
|
- 要素
要素
| #1: タンパク質 | 分子量: 15099.380 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: BRD4, HUNK1 / プラスミド: pNIC28-Bsa4 / 発現宿主: Homo sapiens (ヒト) / 遺伝子: BRD4, HUNK1 / プラスミド: pNIC28-Bsa4 / 発現宿主:  |
|---|---|
| #2: タンパク質・ペプチド | 分子量: 1266.471 Da / 分子数: 1 / 由来タイプ: 合成 詳細: GATA1 peptide acetylated at K312 and K315 C-terminal TYR added for UV detection 由来: (合成)  Homo sapiens (ヒト) / 参照: UniProt: P15976 Homo sapiens (ヒト) / 参照: UniProt: P15976 |
| #3: 化合物 | ChemComp-EDO / |
| #4: 水 | ChemComp-HOH / |
| Has protein modification | Y |
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 2.21 Å3/Da / 溶媒含有率: 44.24 % |
|---|---|
| 結晶化 | 温度: 277 K / 手法: 蒸気拡散法, シッティングドロップ法 / pH: 8 / 詳細: 30.0% PEG 1k 0.1M SPG pH 8.0 |
-データ収集
| 回折 | 平均測定温度: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  Diamond Diamond  / ビームライン: I02 / 波長: 0.9795 Å / ビームライン: I02 / 波長: 0.9795 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 検出器 | タイプ: DECTRIS PILATUS 6M-F / 検出器: PIXEL / 日付: 2016年4月16日 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 放射 | プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 放射波長 | 波長: 0.9795 Å / 相対比: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 反射 | 解像度: 1.4→43.855 Å / Num. all: 28201 / Num. obs: 28201 / % possible obs: 98.4 % / 冗長度: 6.4 % / Rpim(I) all: 0.03 / Rrim(I) all: 0.078 / Rsym value: 0.071 / Net I/av σ(I): 5.7 / Net I/σ(I): 24.4 / Num. measured all: 179422 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 反射 シェル | Diffraction-ID: 1
|
-位相決定
| 位相決定 | 手法:  分子置換 分子置換 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- 解析
解析
| ソフトウェア |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法:  分子置換 分子置換開始モデル: Ensemble of 2OSS, 2OUO, 2GRC, 2OO1, 3DAI, 3D7C, 3DWY 解像度: 1.4→41.17 Å / Cor.coef. Fo:Fc: 0.94 / Cor.coef. Fo:Fc free: 0.923 / SU B: 1.231 / SU ML: 0.023 / SU R Cruickshank DPI: 0.0625 / 交差検証法: THROUGHOUT / σ(F): 0 / ESU R: 0.063 / ESU R Free: 0.058 詳細: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | イオンプローブ半径: 0.8 Å / 減衰半径: 0.8 Å / VDWプローブ半径: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso max: 75.43 Å2 / Biso mean: 9.027 Å2 / Biso min: 2.14 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: final / 解像度: 1.4→41.17 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 拘束条件 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル | 解像度: 1.4→1.436 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj



