[English] 日本語
 Yorodumi
Yorodumi- PDB-6ff3: Crystal structure of Drosophila neural ectodermal development fac... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ff3 | ||||||
|---|---|---|---|---|---|---|---|
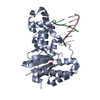
| Title | Crystal structure of Drosophila neural ectodermal development factor Imp-L1 with Human IGF-I | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PEPTIDE BINDING PROTEIN / IGF-I / insulin binding protein / Drosophila / imaginal morphogenesis | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of entry into reproductive diapause / response to insect / glycolate metabolic process / muscle hypertrophy / negative regulation of oocyte development / insulin-like growth factor binding protein complex / insulin-like growth factor ternary complex / positive regulation of trophectodermal cell proliferation / positive regulation of type B pancreatic cell proliferation / prostate gland stromal morphogenesis ...positive regulation of entry into reproductive diapause / response to insect / glycolate metabolic process / muscle hypertrophy / negative regulation of oocyte development / insulin-like growth factor binding protein complex / insulin-like growth factor ternary complex / positive regulation of trophectodermal cell proliferation / positive regulation of type B pancreatic cell proliferation / prostate gland stromal morphogenesis / type II pneumocyte differentiation / neuronal dense core vesicle lumen / positive regulation of glycoprotein biosynthetic process / proteoglycan biosynthetic process / regulation of establishment or maintenance of cell polarity / chondroitin sulfate proteoglycan biosynthetic process / myotube cell development / positive regulation of transcription regulatory region DNA binding / dendrite self-avoidance / negative regulation of neuroinflammatory response / Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) / skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration / bone mineralization involved in bone maturation / IRS-related events triggered by IGF1R / positive regulation of cell growth involved in cardiac muscle cell development / negative regulation of vascular associated smooth muscle cell apoptotic process / positive regulation of cerebellar granule cell precursor proliferation / cell-cell adhesion mediator activity / lung vasculature development / exocytic vesicle / cerebellar granule cell precursor proliferation / positive regulation of myoblast proliferation / lung lobe morphogenesis / positive regulation of myelination / cell activation / negative regulation of androgen receptor signaling pathway / glial cell differentiation / positive regulation of calcineurin-NFAT signaling cascade / transmembrane receptor protein tyrosine kinase activator activity / prostate gland growth / type B pancreatic cell proliferation / mammary gland development / exocrine pancreas development / alphav-beta3 integrin-IGF-1-IGF1R complex / myoblast differentiation / cell surface receptor signaling pathway via STAT / regulation of nitric oxide biosynthetic process / positive regulation of insulin-like growth factor receptor signaling pathway / positive regulation of Ras protein signal transduction / activation of protein kinase B activity / positive regulation of smooth muscle cell migration / growth hormone receptor signaling pathway / insulin binding / positive regulation of DNA binding / negative regulation of interleukin-1 beta production / lung alveolus development / muscle organ development / response to starvation / cellular response to insulin-like growth factor stimulus / branching morphogenesis of an epithelial tube / androgen receptor signaling pathway / positive regulation of cardiac muscle hypertrophy / prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis / negative regulation of release of cytochrome c from mitochondria / type I pneumocyte differentiation / homophilic cell-cell adhesion / negative regulation of amyloid-beta formation / negative regulation of smooth muscle cell apoptotic process / inner ear development / myoblast proliferation / positive regulation of activated T cell proliferation / negative regulation of lipid storage / epithelial to mesenchymal transition / negative regulation of tumor necrosis factor production / Synthesis, secretion, and deacylation of Ghrelin / blood vessel remodeling / positive regulation of glycogen biosynthetic process / positive regulation of insulin receptor signaling pathway / positive regulation of osteoblast differentiation / SHC-related events triggered by IGF1R / postsynaptic modulation of chemical synaptic transmission / positive regulation of vascular associated smooth muscle cell proliferation / insulin-like growth factor receptor binding / extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of mitotic nuclear division / positive regulation of smooth muscle cell proliferation / negative regulation of insulin receptor signaling pathway / insulin-like growth factor receptor signaling pathway / axon guidance / platelet alpha granule lumen / positive regulation of glycolytic process / positive regulation of epithelial cell proliferation / positive regulation of D-glucose import across plasma membrane / determination of adult lifespan / positive regulation of protein secretion / negative regulation of extrinsic apoptotic signaling pathway / insulin receptor binding / skeletal system development / growth factor activity / phosphatidylinositol 3-kinase/protein kinase B signal transduction Similarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.57 Å MOLECULAR REPLACEMENT / Resolution: 2.57 Å | ||||||
 Authors Authors | Brzozowski, A.M. / Kulahin, N. / Kristensen, O. / Schluckebier, G. / Meyts, P.D. / Viola, C.M. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones. Authors: Nikolaj Kulahin Roed / Cristina M Viola / Ole Kristensen / Gerd Schluckebier / Mathias Norrman / Waseem Sajid / John D Wade / Asser Sloth Andersen / Claus Kristensen / Timothy R Ganderton / ...Authors: Nikolaj Kulahin Roed / Cristina M Viola / Ole Kristensen / Gerd Schluckebier / Mathias Norrman / Waseem Sajid / John D Wade / Asser Sloth Andersen / Claus Kristensen / Timothy R Ganderton / Johan P Turkenburg / Pierre De Meyts / Andrzej M Brzozowski /     Abstract: The insulin/insulin-like growth factor signalling axis is an evolutionary ancient and highly conserved hormonal system involved in the regulation of metabolism, growth and lifespan in animals. Human ...The insulin/insulin-like growth factor signalling axis is an evolutionary ancient and highly conserved hormonal system involved in the regulation of metabolism, growth and lifespan in animals. Human insulin is stored in the pancreas, while insulin-like growth factor-1 (IGF-1) is maintained in blood in complexes with IGF-binding proteins (IGFBP1-6). Insect insulin-like polypeptide binding proteins (IBPs) have been considered as IGFBP-like structural and functional homologues. Here, we report structures of the Drosophila IBP Imp-L2 in its free form and bound to Drosophila insulin-like peptide 5 and human IGF-1. Imp-L2 contains two immunoglobulin-like fold domains and its architecture is unrelated to human IGFBPs, suggesting a distinct strategy for bioavailability regulation of insulin-like hormones. Similar hormone binding modes may exist in other insect vectors, as the IBP sequences are highly conserved. Therefore, these findings may open research routes towards a rational interference of transmission of diseases such as malaria, dengue and yellow fevers. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ff3.cif.gz 6ff3.cif.gz | 64 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ff3.ent.gz pdb6ff3.ent.gz | 44.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ff3.json.gz 6ff3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ff/6ff3 https://data.pdbj.org/pub/pdb/validation_reports/ff/6ff3 ftp://data.pdbj.org/pub/pdb/validation_reports/ff/6ff3 ftp://data.pdbj.org/pub/pdb/validation_reports/ff/6ff3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6feyC  4cbpS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 27290.662 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 7663.752 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: IGF1, IBP1 / Production host: Homo sapiens (human) / Gene: IGF1, IBP1 / Production host:  |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.72 Å3/Da / Density % sol: 54.7 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 7.5 Details: cp 10 mg/ml, 4-8% w/v PEG 6K, 5-20 mM MgCl2, 5 mM SB12, 0.1 M TRIS pH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 0.97625 Å / Beamline: I03 / Wavelength: 0.97625 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Feb 28, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97625 Å / Relative weight: 1 |
| Reflection | Resolution: 2.57→48.16 Å / Num. obs: 10666 / % possible obs: 96.4 % / Redundancy: 3.9 % / Rmerge(I) obs: 0.093 / Rpim(I) all: 0.059 / Net I/σ(I): 3.9 |
| Reflection shell | Resolution: 2.57→2.64 Å / Redundancy: 4.1 % / Rmerge(I) obs: 0.783 / Rpim(I) all: 0.47 / % possible all: 97.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4CBP Resolution: 2.57→48.16 Å / Cor.coef. Fo:Fc: 0.932 / Cor.coef. Fo:Fc free: 0.872 / SU B: 15.075 / SU ML: 0.315 / Cross valid method: THROUGHOUT / ESU R: 0.479 / ESU R Free: 0.319 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 58.991 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.57→48.16 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj












